The NMR2 Method to Determine Rapidly the Structure of the Binding Pocket of a Protein–Ligand Complex with High Accuracy
Abstract
:1. Introduction
1.1. Structure-Based Drug Design
1.2. The NMR2 Method
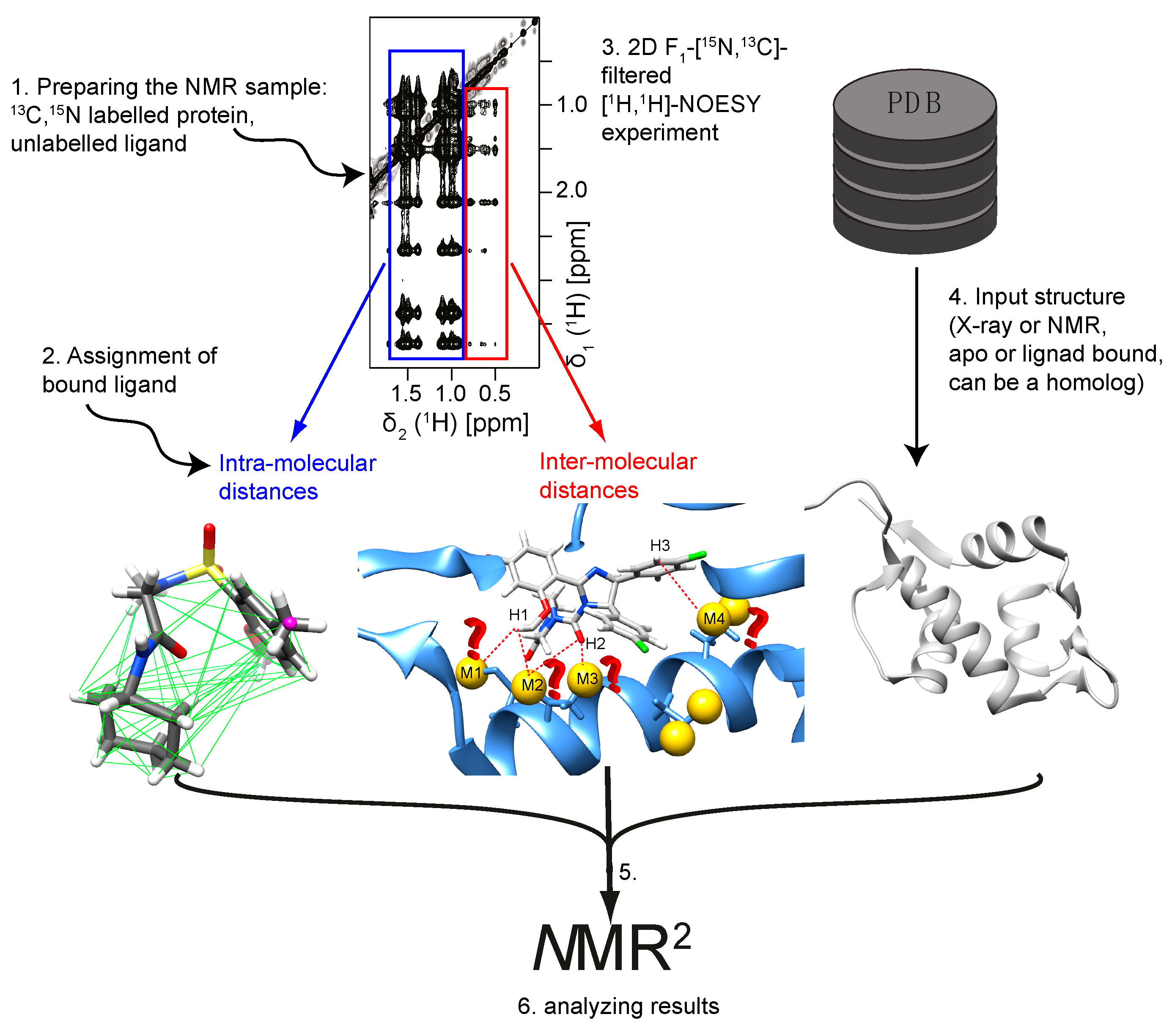
1.3. The NMR2 Protocol
- (i)
- sample preparation for NMR measurements: uniformly 13C,15N labeled, or selective labeling schemes (e.g., isoleucine, leucine, and valine methyl labeling) can be used for the protein [2]. This can be achieved by recombinant expression, e.g., in E. coli [3]. Only one of the two molecules in the complex should be isotopically labeled. For strong binders, i.e., low µM and higher affinity (koff < ΔCS and koff < σ, where koff represents the dissociation rate, ΔCS the chemical shift difference of the bound and free states, and σ the cross-relaxation rate), an equimolar ligand to protein ratio is optimal; whereas for weak binders, i.e., high µM and lower affinity (koff > ΔCS), an excess of ligand is required to saturate the receptor as much as possible. This can be monitored by so-called chemical shift mapping experiments, where the ligand is titrated to the protein and binding is detected through perturbation of the backbone NH chemical shifts of the receptor in 1H,15N-HSQC or TROSY experiments [4,5,6,7]. Knowing the affinity of the small molecule for its receptor, the protein saturation can be calculated with the following formula:where PL, L, P and KD are the concentration of the complex, the concentration of the ligand, the concentration of the protein, and the affinity of the ligand for the protein. The subscript ‘tot’ stands for total concentration.
- (ii)
- Recording experiments to assign the ligand. Usually standard NMR spectra are sufficient to assign the compound in the bound state, e.g., any combination of 13C 1D, 1D DEPT-90, and 1D DEPT-135 spectra [8], 2D 13C,1H-HMQC [9], 2D 13C,1H-HMBC, 2D 1H,1H-DQF COSY [10,11], F1,F2-15N,13C-filtered 1H,1H-TOCSY, or 2D F1,F2-15N,13C-filtered 1H,1H-NOESY spectra [12,13,14,15,16,17,18,19,20,21].
- (iii)
- Measurement of the ligand intra- and ligand–protein inter-molecular distances. All distance restraints for NMR2 are derived from NOE (nuclear Overhauser enhancement) cross-peaks of F1-15N,13C-filtered 1H,1H-NOESY spectra [16,17,18,19,20,21]. These experiments suppress the intra-molecular NOEs peaks from the receptor and render the spectra easier to interpret. In theory, any moiety of the receptor can be analyzed, but to reduce the ambiguity of possible options, the NOEs should be assigned to methyls, amides, or aromatics with respect to their chemical shifts. Focusing only on distinct groups of resonances in the receptor helps to minimize the computational time of the structure calculation. Using methyl groups was so far successful for all complexes. In addition, the NOESY mixing times have to be chosen carefully. The optimal mixing times for the NOE build-ups depend on the correlation time of the complex. Too short of a mixing time would not allow for enough transfer of magnetization and inter-molecular NOE peaks will stay weak or below the noise level. Too long of a mixing time would increase spin diffusion and lead to large signal intensities, but these would require heavy calculations to translate into meaningful distances. In general, NOESY mixing times between 40 and 150 ms are reasonable for a 15–20 kDa protein, exhibiting a correlation time of approximately 10 ns.
- (iv)
- Choosing the input structure. As an input structure, the protein in its apo form, with another bound ligand, or a homolog can be used to derive a starting model of the receptor. Either X-ray or NMR structures can be provided. In the current state of the program, the user should prepare the following input files: a CYANA-regularized protein PDB file, a ligand CYANA library file that can be generated with the program cylib [29], a sequence file containing the amino acid residues of the protein followed by sufficient linker residues (long enough so that the ligand can access all the protein surface) and the ligand residue name as defined in the ligand library file. All these files are needed to produce the starting structure of the complex where the protein structure is identical to the chosen receptor and the ligand is randomly positioned in space but attached to the protein by the linker. Further details can be found in the CYANA manual.
- (v)
- Running NMR2. The NMR2 program screens all possible assignment moieties (usually methyl groups) of the protein and calculates the complex structures for all options. However, it is crucial to diminish the number of options in order to complete the calculations in a reasonable amount of time. This is achieved primarily by using only a fraction of the inter-molecular distances in the first calculation cycle, where only around 3–4 methyl groups of the protein are taken into account. The use of an input structure, the previously derived network of inter-molecular distances, and the use of triangle or tetra angle smoothing to rule out most of the false assignment possibilities are equally important for a manageable calculation time. As of now, NMR2 is a CYANA-based program and calculates all structures using the standard simulated annealing protocol [30]. The results are scored with respect to the target function, which represents a measure of how well the calculated structure fulfills the data. CYANA is the most widely used NMR structure calculation program, which is solely based on experimental data and the repulsive part of the van der Waals potential modeling the atom radii. No other force field is used and therefore the electrostatic potential of the molecules is not modelled. Nonetheless, if specific interactions are known or determined by experiments they can be added following the program syntax [30]. Only the best structures are kept for the next calculation cycle where more methyl groups with their respective inter-molecular distances are included. The calculation is finished when all experimental data have been used.
- (vi)
- Analyzing the results. The final complex structures have to be analyzed carefully to detect potential errors. NMR2 requires a definition of the receptor flexibility; however, if there are no restraints on backbone and side chain atoms, the protein will freely move to fulfill the distance restraints, which could potentially yield false positives. Another source of false positives is when the ligand finds its binding site at the N- or C-terminus of the protein or where the protein atom density is the lowest. There, the ligand can freely adopt its position and orientation to fulfill the distance restraints because little or no steric inter-molecular interactions are present. One should keep in mind that this is happening only if the protein contains methyl groups at these sites.
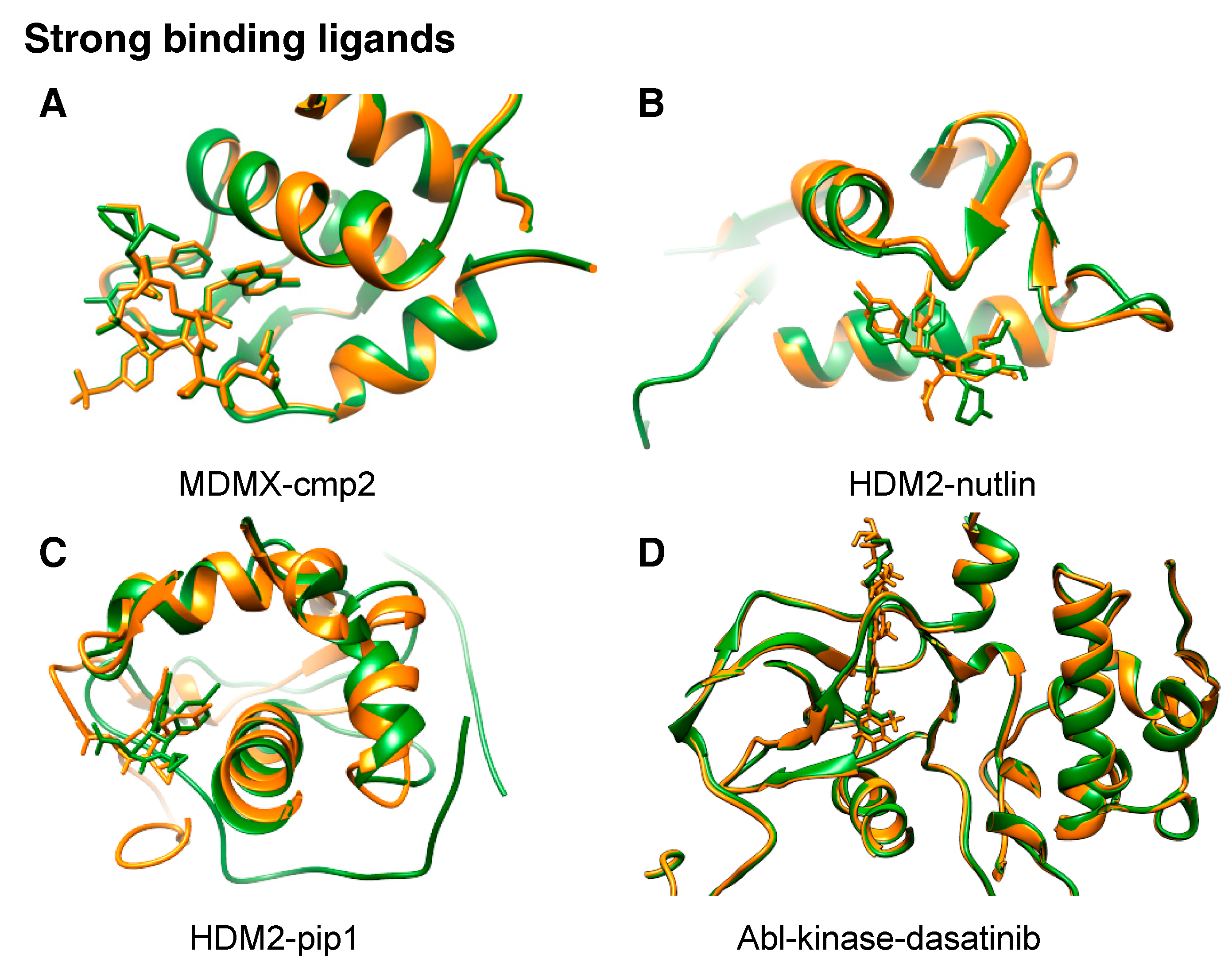
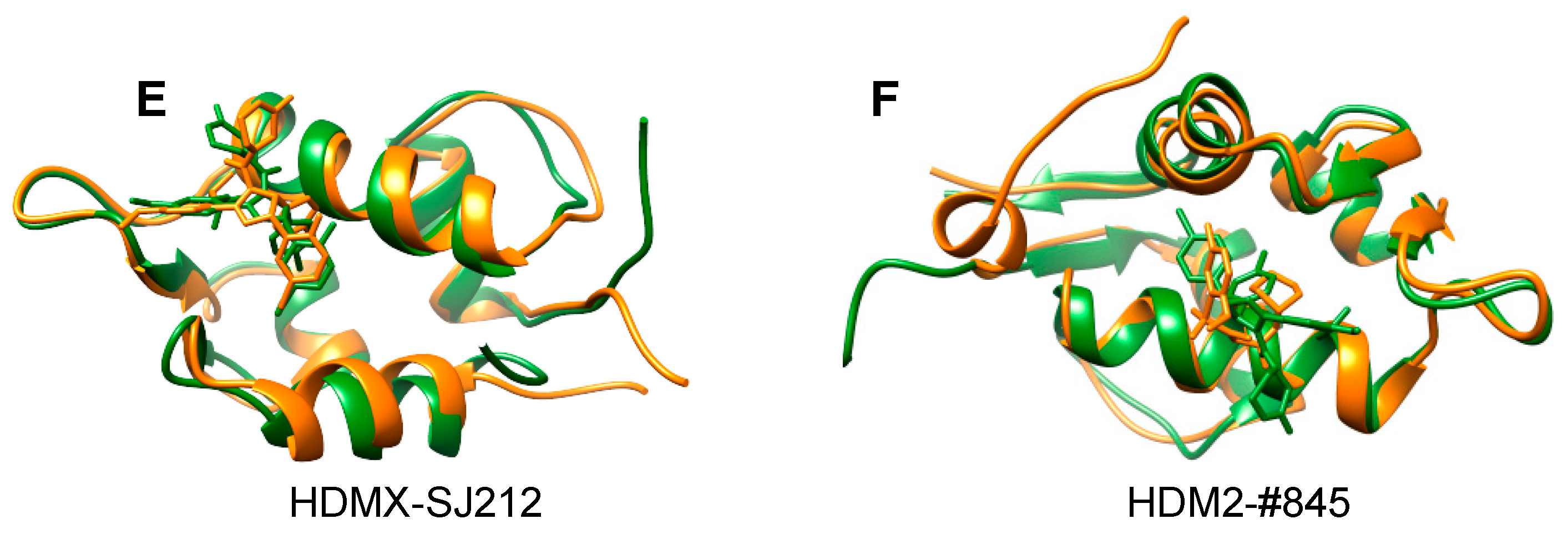
1.4. Current Applications of NMR2
1.5. NMR2 versus Other Methods for Rapid Structure Calculations of Protein–Ligand Complexes
2. Conclusions and Outlook
Acknowledgments
Conflicts of Interest
References
- Drews, J. Drug discovery: A historical perspective. Science 2000, 287, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Tugarinov, V.; Kay, L.E. Methyl groups as probes of structure and dynamics in nmr studies of high-molecular-weight proteins. Chembiochem 2005, 6, 1567–1577. [Google Scholar] [CrossRef] [PubMed]
- Mcintosh, L.P.; Dahlquist, F.W. Biosynthetic incorporation of n-15 and c-13 for assignment and interpretation of nuclear-magnetic-resonance spectra of proteins. Q. Rev. Biophys. 1990, 23, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Pervushin, K.; Riek, R.; Wider, G.; Wuthrich, K. Attenuated t-2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to nmr structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 1997, 94, 12366–12371. [Google Scholar] [CrossRef] [PubMed]
- Ziarek, J.J.; Peterson, F.C.; Lytle, B.L.; Volkman, B.F. Binding site identification and structure determination of protein-ligand complexes by nmr: A semiautomated approach. Methods Enzymol. 2011, 493, 241–275. [Google Scholar] [PubMed]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Mag. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zuiderweg, E.R.P. Mapping protein-protein interactions in solution by nmr spectroscopy. Biochemistry 2002, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bendall, M.R.; Doddrell, D.M.; Pegg, D.T. Editing of c-13 nmr-spectra—A pulse sequence for the generation of subspectra. J. Am. Chem. Soc. 1981, 103, 4603–4605. [Google Scholar] [CrossRef]
- Bax, A.; Griffey, R.H.; Hawkins, B.L. Correlation of proton and n-15 chemical-shifts by multiple quantum nmr. J. Magn. Reson. 1983, 55, 301–315. [Google Scholar] [CrossRef]
- Aue, W.P.; Bartholdi, E.; Ernst, R.R. 2-dimensional spectroscopy—Application to nuclear magnetic-resonance. J. Chem. Phys. 1976, 64, 2229–2246. [Google Scholar] [CrossRef]
- Piantini, U.; Sorensen, O.W.; Ernst, R.R. Multiple quantum filters for elucidating nmr coupling networks. J. Am. Chem. Soc. 1982, 104, 6800–6801. [Google Scholar] [CrossRef]
- Ni, F. Complete relaxation matrix analysis of transferred nuclear overhauser effects. J. Magn. Reson. 1992, 96, 651–656. [Google Scholar] [CrossRef]
- Ni, F. Recent developments in transferred noe methods. Prog. Nucl. Magn. Reson. Spectrosc. 1994, 26, 517–606. [Google Scholar] [CrossRef]
- Ni, F.; Zhu, Y. Accounting for ligand-protein interactions in the relaxation-matrix analysis of transferred nuclear overhauser effects. J. Magn. Reson. Ser. B 1994, 103, 180–184. [Google Scholar] [CrossRef]
- Berger, S.; Braun, S. 200 and More Nmr Experiments: A Practical Course; Wiley: Weinheim, Germany, 2004. [Google Scholar]
- Breeze, A.L. Isotope-filtered nmr methods for the study of biomolecular structure and interactions. Prog. Nucl. Magn. Reson. Spectrosc. 2000, 36, 323–372. [Google Scholar] [CrossRef]
- Iwahara, J.; Wojciak, J.M.; Clubb, R.T. Improved nmr spectra of a protein-DNA complex through rational mutagenesis and the application of a sensitivity optimized isotope-filtered noesy experiment. J. Biomol. NMR 2001, 19, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Kogler, H.; Sorensen, O.W.; Bodenhausen, G.; Ernst, R.R. Low-pass j-filters—Suppression of neighbor peaks in heteronuclear relayed correlation spectra. J. Magn. Reson. 1983, 55, 157–163. [Google Scholar] [CrossRef]
- Ogura, K.; Terasawa, H.; Inagaki, F. An improved double-tuned and isotope-filtered pulse scheme based on a pulsed field gradient and a wide-band inversion shaped pulse. J. Biomol. NMR 1996, 8, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Otting, G.; Wuthrich, K. Heteronuclear filters in 2-dimensional [h-1, h-1] nmr-spectroscopy—Combined use with isotope labeling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys. 1990, 23, 39–96. [Google Scholar] [CrossRef] [PubMed]
- Zwahlen, C.; Legault, P.; Vincent, S.J.F.; Greenblatt, J.; Konrat, R.; Kay, L.E. Methods for measurement of intermolecular noes by multinuclear nmr spectroscopy: Application to a bacteriophage lambda n-peptide/boxb rna complex. J. Am. Chem. Soc. 1997, 119, 6711–6721. [Google Scholar] [CrossRef]
- Macura, S.; Ernst, R.R. Elucidation of cross relaxation in liquids by two-dimensional nmr-spectroscopy. Mol. Phys. 1980, 41, 95–117. [Google Scholar] [CrossRef]
- Solomon, I. Relaxation processes in a system of 2 spins. Phys. Rev. 1955, 99, 559–565. [Google Scholar] [CrossRef]
- Orts, J.; Vogeli, B.; Riek, R. Relaxation matrix analysis of spin diffusion for the nmr structure calculation with enoes. J. Chem. Theory Comput. 2012, 8, 3483–3492. [Google Scholar] [CrossRef] [PubMed]
- Strotz, D.; Orts, J.; Chi, C.N.; Riek, R.; Vogeli, B. Enora2 exact noe analysis program. J. Chem. Theory Comput. 2017, 13, 4336–4346. [Google Scholar] [CrossRef] [PubMed]
- Reuben, J.; Fiat, D. Nuclear magnetic resonance studies of solutions of rare-earth ions and their complexes. 4. Concentration and temperature dependence of oxygen-14 transverse relaxation in aqueous solutions. J. Chem. Phys. 1969, 51, 4918–4927. [Google Scholar] [CrossRef]
- Lippens, G.M.; Cerf, C.; Hallenga, K. Theory and experimental results of transfer-noe experiments. 1. The influence of the off rate versus cross-relaxation rates. J. Magn. Reson. 1992, 99, 268–281. [Google Scholar] [CrossRef]
- Cavanagh, J.; Fairbrother, W.J.; Palmer, A.G.; Rance, M.; Skelton, N.J. Protein nmr spectroscopy principles and practice second edition preface. In Protein Nmr Spectroscopy: Principles and Practice, 2nd ed.; Academic Press: Burlington, MA, USA, 2007; pp. V–VI. [Google Scholar]
- Yilmaz, E.M.; Guntert, P. Nmr structure calculation for all small molecule ligands and non-standard residues from the pdb chemical component dictionary. J. Biomol. NMR 2015, 63, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Guntert, P.; Mumenthaler, C.; Wuthrich, K. Torsion angle dynamics for nmr structure calculation with the new program dyana. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.L.; Mao, X.A.; Ye, C.H.; Huang, H.; Nicholson, J.K.; Lindon, J.C. Improved watergate pulse sequences for solvent suppression in nmr spectroscopy. J. Magn. Reson. 1998, 132, 125–129. [Google Scholar] [CrossRef]
- Orts, J.; Walti, M.A.; Marsh, M.; Vera, L.; Gossert, A.D.; Guntert, P.; Riek, R. Nmr-based determination of the 3d structure of the ligand-protein interaction site without protein resonance assignment. J. Am. Chem. Soc. 2016, 138, 4393–4400. [Google Scholar] [CrossRef] [PubMed]
- Walti, M.A.; Riek, R.; Orts, J. Fast nmr-based determination of the 3d structure of the binding site of protein-ligand complexes with weak affinity binders. Angew. Chem. Int. Ed. 2017, 56, 5208–5211. [Google Scholar] [CrossRef] [PubMed]
- Kallen, J.; Goepfert, A.; Blechschmidt, A.; Izaac, A.; Geiser, M.; Tavares, G.; Ramage, P.; Furet, P.; Masuya, K.; Lisztwan, J. Crystal structures of human mdmx (hdmx) in complex with p53 peptide analogues reveal surprising conformational changes. J. Biol. Chem. 2009, 284, 8803–8812. [Google Scholar] [CrossRef] [PubMed]
- Michelsen, K.; Jordan, J.B.; Lewis, J.; Long, A.M.; Yang, E.; Rew, Y.; Zhou, J.; Yakowec, P.; Schnier, P.D.; Huang, X.; et al. Ordering of the n-terminus of human mdm2 by small molecule inhibitors. J. Am. Chem. Soc. 2012, 134, 17059–17067. [Google Scholar] [CrossRef] [PubMed]
- Tokarski, J.S.; Newitt, J.A.; Chang, C.Y.J.; Cheng, J.D.; Wittekind, M.; Kiefer, S.E.; Kish, K.; Lee, F.Y.F.; Borzillerri, R.; Lombardo, L.J.; et al. The structure of dasatinib (bms-354825) bound to activated abl kinase domain elucidates its inhibitory activity against imatinib-resistant abl mutants. Cancer Res. 2006, 66, 5790–5797. [Google Scholar] [CrossRef] [PubMed]
- Grace, C.R.; Ban, D.; Min, J.; Mayasundari, A.; Min, L.; Finch, K.E.; Griffiths, L.; Bharatham, N.; Bashford, D.; Guy, R.K.; et al. Monitoring ligand-induced protein ordering in drug discovery. J. Mol. Biol. 2016, 428, 1290–1303. [Google Scholar] [CrossRef] [PubMed]
- Nilges, M.; Macias, M.J.; ODonoghue, S.I.; Oschkinat, H. Automated noesy interpretation with ambiguous distance restraints: The refined nmr solution structure of the pleckstrin homology domain from beta-spectrin. J. Mol. Biol. 1997, 269, 408–422. [Google Scholar] [CrossRef] [PubMed]
- Nilges, M.; O’Donoghue, S.I. Ambiguous noes and automated noe assignment. Prog. Nucl. Magn. Reson. Spectrosc. 1998, 32, 107–139. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. Haddock: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Clore, G.M.; Schwieters, C.D. Docking of protein-protein complexes on the basis of highly ambiguous intermolecular distance restraints derived from h-1(n)/n-15 chemical shift mapping and backbone n-15-h-1 residual dipolar couplings using conjoined rigid body/torsion angle dynamics. J. Am. Chem. Soc. 2003, 125, 2902–2912. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Westerhoff, L.M.; Merz, K.M. A critical assessment of the performance of protein-ligand scoring functions based on nmr chemical shift perturbations. J. Med. Chem. 2007, 50, 5128–5134. [Google Scholar] [CrossRef] [PubMed]
- McCoy, M.A.; Wyss, D.F. Spatial localization of ligand binding sites from electron current density surfaces calculated from nmr chemical shift perturbations. J. Am. Chem. Soc. 2002, 124, 11758–11763. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.; Hunter, C.A.; Packer, M.J.; Spitaleri, A. Determination of protein-ligand binding modes using complexation-induced changes in h-1 nmr chemical shift. J. Med. Chem. 2008, 51, 2512–2517. [Google Scholar] [CrossRef] [PubMed]
- Hajduk, P.J.; Mack, J.C.; Olejniczak, E.T.; Park, C.; Dandliker, P.J.; Beutel, B.A. Sos-nmr: A saturation transfer nmr-based method for determining the structures of protein-ligand complexes. J. Am. Chem. Soc. 2004, 126, 2390–2398. [Google Scholar] [CrossRef] [PubMed]
- Jayalakshmi, V.; Krishna, N.R. Corcema refinement of the bound ligand conformation within the protein binding pocket in reversibly forming weak complexes using std-nmr intensities. J. Magn. Reson. 2004, 168, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Schieborr, U.; Vogtherr, M.; Elshorst, B.; Betz, M.; Grimme, S.; Pescatore, B.; Langer, T.; Saxena, K.; Schwalbe, H. How much nmr data is required to determine a protein-ligand complex structure? Chembiochem 2005, 6, 1891–1898. [Google Scholar] [CrossRef] [PubMed]
- Orts, J.; Bartoschek, S.; Griesinger, C.; Monecke, P.; Carlomagno, T. An nmr-based scoring function improves the accuracy of binding pose predictions by docking by two orders of magnitude. J. Biomol. NMR 2012, 52, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Constantine, K.L.; Davis, M.E.; Metzler, W.J.; Mueller, L.; Claus, B.L. Protein-ligand noe matching: A high-throughput method for binding pose evaluation that does not require protein nmr resonance assignments. J. Am. Chem. Soc. 2006, 128, 7252–7263. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Sykes, B.D. The c-13 chemical-shift index—A simple method for the identification of protein secondary structure using c-13 chemical-shift data. J. Biomol. NMR 1994, 4, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Williamson, M.P.; Kikuchi, J.; Asakura, T. Application of h-1-nmr chemical-shifts to measure the quality of protein structures. J. Mol. Biol. 1995, 247, 541–546. [Google Scholar] [CrossRef]
- Wishart, D.S.; Watson, M.S.; Boyko, R.F.; Sykes, B.D. Automated h-1 and c-13 chemical shift prediction using the biomagresbank. J. Biomol. NMR 1997, 10, 329–336. [Google Scholar] [CrossRef] [PubMed]
- McCoy, M.A.; Wyss, D.F. Alignment of weakly interacting molecules to protein surfaces using simulations of chemical shift perturbations. J. Biomol. NMR 2000, 18, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.; Hunter, C.A.; Packer, M.J.; Pandya, M.J.; Williamson, M.P. Use of quantitative (1)h nmr chemical shift changes for ligand docking into barnase. J. Biomol. NMR 2009, 43, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Shuker, S.B.; Hajduk, P.J.; Meadows, R.P.; Fesik, S.W. Discovering high-affinity ligands for proteins: Sar by nmr. Science 1996, 274, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Farmer, B.T. Localizing the nadp(+) binding site on the murb enzyme by nmr. Nat. Struct. Biol. 1996, 3, 995–997. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, P.; Waygood, E.B.; Reizer, J.; Saier, M.H.; Klevit, R.E. Demonstration of protein-protein interaction specificity by nmr chemical shift mapping. Protein Sci. 1997, 6, 2624–2627. [Google Scholar] [CrossRef] [PubMed]
- Schmiedeskamp, M.; Rajagopal, P.; Klevit, R.E. Nmr chemical shift perturbation mapping of DNA binding by a zinc-finger domain from the yeast transcription factor adr1. Protein Sci. 1997, 6, 1835–1848. [Google Scholar] [CrossRef] [PubMed]
- McCoy, M.A.; Wyss, D.F. Structures of protein-protein complexes are docked using only nmr restraints from residual dipolar coupling and chemical shift perturbations. J. Am. Chem. Soc. 2002, 124, 2104–2105. [Google Scholar] [CrossRef] [PubMed]
- Trellet, M.; Melquiond, A.S.J.; Bonvin, A.M.J.J. A unified conformational selection and induced fit approach to protein-peptide docking. PLoS ONE 2013, 8, e58769. [Google Scholar] [CrossRef] [PubMed]
- Arnesano, F.; Banci, L.; Piccioli, M. Nmr structures of paramagnetic metalloproteins. Q. Rev. Biophys. 2005, 38, 167–219. [Google Scholar] [CrossRef] [PubMed]
- Otting, G. Prospects for lanthanides in structural biology by nmr. J. Biomol. NMR 2008, 42, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Palma, P.N.; Krippahl, L.; Wampler, J.E.; Moura, J.J.G. Bigger: A new (soft) docking algorithm for predicting protein interactions. Proteins 2000, 39, 372–384. [Google Scholar] [CrossRef]
- Stark, J.; Powers, R. Rapid protein-ligand costructures using chemical shift perturbations. J. Am. Chem. Soc. 2008, 130, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Krzeminski, M.; Loth, K.; Boelens, R.; Bonvin, A.M.J.J. Samplex: Automatic mapping of perturbed and unperturbed regions of proteins and complexes. BMC Bioinform. 2010, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pedregal, V.M.; Reese, M.; Meiler, J.; Blommers, M.J.J.; Griesinger, C.; Carlomagno, T. The inpharma method: Protein-mediated interligand noes for pharmacophore mapping. Angew. Chem. Int. Ed. 2005, 44, 4172–4175. [Google Scholar] [CrossRef] [PubMed]
- Stauch, B.; Orts, J.; Carlomagno, T. The description of protein internal motions aids selection of ligand binding poses by the inpharma method. J. Biomol. NMR 2012, 54, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Curto, E.V.; Moseley, H.N.B.; Krishna, N.R. Corcema evaluation of the potential role of intermolecular transferred noesy in the characterization of ligand-receptor complexes. J. Comput. Aided Mol. Des. 1996, 10, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Moseley, H.N.B.; Curto, E.V.; Krishna, N.R. Complete relaxation and conformational exchange matrix (corcema) analysis of noesy spectra of interacting systems—2-dimensional transferred noesy. J. Magn. Reson. Ser. B 1995, 108, 243–261. [Google Scholar] [CrossRef]
- Kuntz, I.D.; Blaney, J.M.; Oatley, S.J.; Langridge, R.; Ferrin, T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982, 161, 269–288. [Google Scholar] [CrossRef]
- Kuntz, I.D.; Meng, E.C.; Shoichet, B.K. Structure-based molecular design. Accounts Chem. Res. 1994, 27, 117–123. [Google Scholar] [CrossRef]
- Orts, J.; Grimm, S.K.; Griesinger, C.; Wendt, K.U.; Bartoschek, S.; Carlomagno, T. Specific methyl group protonation for the measurement of pharmacophore-specific interligand noe interactions. Chem. Eur. J. 2008, 14, 7517–7520. [Google Scholar] [CrossRef] [PubMed]
- Orts, J.; Tuma, J.; Reese, M.; Grimm, S.K.; Monecke, P.; Bartoschek, S.; Schiffer, A.; Wendt, K.U.; Griesinger, C.; Carlomagno, T. Crystallography-independent determination of ligand binding modes. Angew. Chem. Int. Ed. 2008, 47, 7736–7740. [Google Scholar] [CrossRef] [PubMed]
- Vogeli, B.; Segawa, T.F.; Leitz, D.; Sobol, A.; Choutko, A.; Trzesniak, D.; van Gunsteren, W.; Riek, R. Exact distances and internal dynamics of perdeuterated ubiquitin from noe buildups. J. Am. Chem. Soc. 2009, 131, 17215–17225. [Google Scholar] [CrossRef] [PubMed]
- Vogeli, B.; Orts, J.; Strotz, D.; Chi, C.; Minges, M.; Walti, M.A.; Guntert, P.; Riek, R. Towards a true protein movie: A perspective on the potential impact of the ensemble-based structure determination using exact noes. J. Magn. Reson. 2014, 241, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.G.; Blow, D.M. Detection of sub-units within crystallographic asymmetric unit. Acta Crystallogr. 1962, 15, 24–31. [Google Scholar] [CrossRef]
- Hillisch, A.; Pineda, L.F.; Hilgenfeld, R. Utility of homology models in the drug discovery process. Drug Discov. Today 2004, 9, 659–669. [Google Scholar] [CrossRef]
- Schirmer, R.E.; Noggle, J.H. Quantitative application of nuclear overhauser effect to determination of molecular structure. J. Am. Chem. Soc. 1972, 94, 2947–2952. [Google Scholar] [CrossRef]
- Balaram, P.; Bothnerb, A.; Breslow, E. Localization of tyrosine at binding-site of neurophysin ii by negative nuclear overhauser effects. J. Am. Chem. Soc. 1972, 94, 4017–4018. [Google Scholar] [CrossRef]
- Kaiser, R. Intermolecular nuclear overhauser effect in liquid solutions. J. Chem. Phys. 1965, 42, 1838–1839. [Google Scholar] [CrossRef]
- Pritisanac, I.; Degiacomi, M.T.; Alderson, T.R.; Carneiro, M.G.; Eiso, A.B.; Siegal, G.; Baldwin, A.J. Automatic assignment of methyl-nmr spectra of supramolecular machines using graph theory. J. Am. Chem. Soc. 2017, 139, 9523–9533. [Google Scholar] [CrossRef] [PubMed]
- Chao, F.A.; Kim, J.; Xia, Y.; Milligan, M.; Rowe, N.; Veglia, G. Flamengo 2.0: An enhanced fuzzy logic algorithm for structure-based assignment of methyl group resonances. J. Magn. Reson. 2014, 245, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Matthews, S. Map-xsii: An improved program for the automatic assignment of methyl resonances in large proteins. J. Biomol. NMR 2013, 55, 179–187. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wälti, M.A.; Orts, J. The NMR2 Method to Determine Rapidly the Structure of the Binding Pocket of a Protein–Ligand Complex with High Accuracy. Magnetochemistry 2018, 4, 12. https://doi.org/10.3390/magnetochemistry4010012
Wälti MA, Orts J. The NMR2 Method to Determine Rapidly the Structure of the Binding Pocket of a Protein–Ligand Complex with High Accuracy. Magnetochemistry. 2018; 4(1):12. https://doi.org/10.3390/magnetochemistry4010012
Chicago/Turabian StyleWälti, Marielle Aulikki, and Julien Orts. 2018. "The NMR2 Method to Determine Rapidly the Structure of the Binding Pocket of a Protein–Ligand Complex with High Accuracy" Magnetochemistry 4, no. 1: 12. https://doi.org/10.3390/magnetochemistry4010012




