Precision and Accuracy Limits of Wastewater-Based Epidemiology—Lessons Learned from SARS-CoV-2: A Scoping Review
Abstract
:1. Introduction
2. Materials and Methods
3. Results
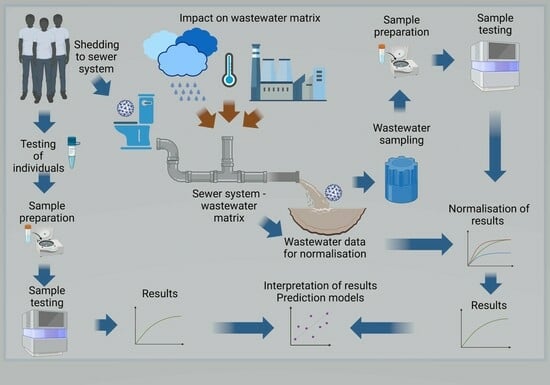
3.1. The Wastewater-Based Epidemiology Concept
3.2. SARS-CoV-2 RNA Detection: Risks and Data Interpretation
3.3. Wastewater Surveillance and Early Warning Systems in Communities
3.4. Shedding Sources and Rates
3.5. Infectivity of the Detected RNA
3.6. Wastewater Sampling
3.7. Wastewater Sample Preparation and Analysis
3.8. Sample Normalization
3.9. Wastewater Matrix and Environmental Factors
4. Discussion
- Appropriate normalization methods to determine the most efficient process, especially using wastewater parameters and measurements, which are used in the usual monitoring performance of most WWTPs. These include the specific wastewater amount per person, TSSs, BOD, COD, N and its forms, P and its forms, temperature, pH, conductivity, or a combination.
- Appropriate factor identification in the latter, which has a higher impact on the virus detection, quantification, and estimation of the actual number of infected persons using WBE during virus RNA transport within the sewer system.
- The transposition and interpretation of the socio-economic aspects of the wastewater composition parameters.
- Development of mathematical models for virus, bacteria, or chemical transport within sewerage systems.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Galani, A.; Aalizadeh, R.; Kostakis, M.; Markou, A.; Alygizakis, N.; Lytras, T.; Adamopoulos, P.G.; Peccia, J.; Thompson, D.C.; Kontou, A.; et al. SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 2022, 804, 150151. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Gao, M.; Chen, C.H.; Zhou, Y.; Zhan, Z.H.; Ren, Y. Knowledge graph of wastewater-based epidemiology development: A data-driven analysis based on research topics and trends. Environ. Sci. Pollut. Res. 2023, 30, 28373–28382. [Google Scholar] [CrossRef] [PubMed]
- Faraway, J.; Boxall-Clasby, J.; Feil, E.J.; Gibbon, M.J.; Hatfield, O.; Kasprzyk-Hordern, B.; Smith, T. Challenges in realising the potential of wastewater-based epidemiology to quantitatively monitor and predict the spread of disease. J. Water Health 2022, 20, 1038–1050. [Google Scholar] [CrossRef] [PubMed]
- Hart, O.E.; Halden, R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020, 730, 138875. [Google Scholar] [CrossRef]
- Hou, C.; Chu, T.; Chen, M.; Hua, Z.; Xu, P.; Xu, H.; Wang, Y.; Liao, J.; Di, B. Application of multi-parameter population model based on endogenous population biomarkers and flow volume in wastewater epidemiology. Sci. Total Environ. 2021, 759, 143480. [Google Scholar] [CrossRef] [PubMed]
- Berzina, Z.; Pavlenko, R.; Jansons, M.; Bartkiene, E.; Neilands, R.; Pugajeva, I.; Bartkevics, V. Application of Wastewater-Based Epidemiology for Tracking Human Exposure to Deoxynivalenol and Enniatins. Toxins 2022, 14, 91. [Google Scholar] [CrossRef] [PubMed]
- Tomsone, L.E.; Perkons, I.; Sukajeva, V.; Neilands, R.; Kokina, K.; Bartkevics, V.; Pugajeva, I. Consumption trends of pharmaceuticals and psychoactive drugs in Latvia determined by the analysis of wastewater. Water Res. 2022, 221, 118800. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, B.; Yang, L.; Huo, T.; Bai, D.; An, Q.; Li, X. Consumption of common illicit drugs in twenty-one cities in southwest China through wastewater analysis. Sci. Total Environ. 2022, 851, 158105. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Shi, J.; Luby, S.P.; Jiang, G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021, 415, 129039. [Google Scholar] [CrossRef]
- Polo, D.; Quintela-Baluja, M.; Corbishley, A.; Jones, D.L.; Singer, A.C.; Graham, D.W.; Romalde, J.L. Making waves: Wastewater-based epidemiology for COVID-19—Approaches and challenges for surveillance and prediction. Water Res. 2020, 186, 116404. [Google Scholar] [CrossRef]
- Zhang, D.; Duran, S.S.F.; Lim, W.Y.S.; Tan, C.K.I.; Cheong, W.C.D.; Suwardi, A.; Loh, X.J. SARS-CoV-2 in wastewater: From detection to evaluation. Mater. Today Adv. 2022, 13, 100211. [Google Scholar] [CrossRef] [PubMed]
- Domokos, E.; Sebestyén, V.; Somogyi, V.; Trájer, A.J.; Gerencsér-Berta, R.; Oláhné Horváth, B.; Tóth, E.G.; Jakab, F.; Kemenesi, G.; Abonyi, J. Identification of sampling points for the detection of SARS-CoV-2 in the sewage system. Sustain. Cities Soc. 2022, 76, 103422. [Google Scholar] [CrossRef] [PubMed]
- Haak, L.; Delic, B.; Li, L.; Guarin, T.; Mazurowski, L.; Dastjerdi, N.G.; Dewan, A.; Pagilla, K. Spatial and temporal variability and data bias in wastewater surveillance of SARS-CoV-2 in a sewer system. Sci. Total Environ. 2022, 805, 150390. [Google Scholar] [CrossRef] [PubMed]
- Gudra, D.; Dejus, S.; Bartkevics, V.; Roga, A.; Kalnina, I.; Strods, M.; Rayan, A.; Kokina, K.; Zajakina, A.; Dumpis, U.; et al. Detection of SARS-CoV-2 RNA in wastewater and importance of population size assessment in smaller cities: An exploratory case study from two municipalities in Latvia. Sci. Total Environ. 2022, 823, 153775. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Avilés, P.; Moreno-Andrade, I.; Carrillo-Reyes, J. Approaches applied to detect SARS-CoV-2 in wastewater and perspectives post-COVID-19. J. Water Process Eng. 2021, 40, 101947. [Google Scholar] [CrossRef] [PubMed]
- Mahon, J.M.; Monleon, A.J.C.; Gill, L.W.; O’Sullivan, J.J.; Meijer, W.G. Wastewater-based epidemiology (WBE) for SARS-CoV-2—A review focussing on the significance of the sewer network using a Dublin city catchment case study. Water Sci. Technol. 2022, 86, 1402–1425. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.N.; Le, G.T.; Nguyen, D.T.; Juang, R.S.; Rinklebe, J.; Bhatnagar, A.; Lima, E.C.; Iqbal, H.M.N.; Sarmah, A.K.; Chao, H.P. SARS-CoV-2 coronavirus in water and wastewater: A critical review about presence and concern. Environ. Res. 2021, 193, 110265. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Guidelines and Guidance Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Boogaerts, T.; Ahmed, F.; Choi, P.M.; Tscharke, B.; O’Brien, J.; De Loof, H.; Gao, J.; Thai, P.; Thomas, K.; Mueller, J.F.; et al. Current and future perspectives for wastewater-based epidemiology as a monitoring tool for pharmaceutical use. Sci. Total Environ. 2021, 789, 148047. [Google Scholar] [CrossRef]
- Elsevier. Scopus Data Base. Available online: www.scopus.com (accessed on 26 November 2023).
- Li, X.; Zhang, S.; Sherchan, S.; Orive, G.; Lertxundi, U.; Haramoto, E.; Honda, R.; Kumar, M.; Arora, S.; Kitajima, M. Correlation between SARS-CoV-2 RNA concentration in wastewater and COVID-19 cases in community: A systematic review and meta-analysis. J. Hazard. Mater. 2023, 441, 129848. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kulandaivelu, J.; Guo, Y.; Zhang, S.; Shi, J.; O’Brien, J.; Arora, S.; Kumar, M.; Sherchan, S.P.; Honda, R.; et al. SARS-CoV-2 shedding sources in wastewater and implications for wastewater-based epidemiology. J. Hazard. Mater. 2022, 432, 128667. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Xiao, A.; Zhang, J.; Moniz, K.; Endo, N.; Armas, F.; Bonneau, R.; Brown, M.A.; Bushman, M.; Chai, P.R.; et al. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 2022, 805, 150121. [Google Scholar] [CrossRef] [PubMed]
- Von Sperling, M.; Verbyla, M.E.; Oliveira, S.M.A.C. Relationship between monitoring variables. Correlation and regression analysis. In Assessment of Treatment Plant Performance and Water Quality Data: A Guide for Students, Researchers and Practitioners; IWA Publishing: London, UK, 2020; pp. 397–477. [Google Scholar] [CrossRef]
- Weidhaas, J.; Aanderud, Z.T.; Roper, D.K.; VanDerslice, J.; Gaddis, E.B.; Ostermiller, J.; Hoffman, K.; Jamal, R.; Heck, P.; Zhang, Y.; et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021, 775, 145790. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lee, B.E.; Gao, T.; Qiu, Y.; Ellehoj, E.; Yu, J.; Diggle, M.; Tipples, G.; Maal-Bared, R.; Hinshaw, D.; et al. Number of COVID-19 cases required in a population to detect SARS-CoV-2 RNA in wastewater in the province of Alberta, Canada: Sensitivity assessment. J. Environ. Sci. 2023, 125, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, V.; Al-Duroobi, H.; Phan, D.C.; Palekar, R.S.; Blount, B.; Rambhia, K.J. Wastewater surveillance for SARS-CoV-2 to support return to campus: Methodological considerations and data interpretation. Curr. Opin. Environ. Sci. Health 2022, 27, 100362. [Google Scholar] [CrossRef] [PubMed]
- Adeel, M.; Farooq, T.; Shakoor, N.; Ahmar, S.; Fiaz, S.; White, J.C.; Gardea-Torresdey, J.L.; Mora-Poblete, F.; Rui, Y. COVID-19 and nanoscience in the developing world: Rapid detection and remediation in wastewater. Nanomaterials 2021, 11, 991. [Google Scholar] [CrossRef]
- Assoum, M.; Lau, C.L.; Thai, P.K.; Ahmed, W.; Mueller, J.F.; Thomas, K.V.; Choi, P.M.; Jackson, G.; Selvey, L.A. Wastewater Surveillance Can Function as an Early Warning System for COVID-19 in Low-Incidence Settings. Trop. Med. Infect. Dis. 2023, 8, 211. [Google Scholar] [CrossRef]
- Murni, I.K.; Oktaria, V.; Handley, A.; McCarthy, D.T.; Donato, C.M.; Nuryastuti, T.; Supriyati, E.; Putri, D.A.D.; Sari, H.M.; Laksono, I.S.; et al. The feasibility of SARS-CoV-2 surveillance using wastewater and environmental sampling in Indonesia. PLoS ONE 2022, 17, e274793. [Google Scholar] [CrossRef]
- Keshaviah, A.; Hu, X.C.; Henry, M. Developing a flexible national wastewater surveillance system for COVID-19 and beyond. Environ. Health Perspect. 2021, 129, 045002. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, J.V.; Romero, D.; Kopka, C.J.; Karim, S.A.; Abu-Raddad, L.J.; Almeida, G.; Baptista-Leite, R.; Barocas, J.A.; Barreto, M.L.; Bar-Yam, Y.; et al. A multinational Delphi consensus to end the COVID-19 public health threat. Nature 2022, 611, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Vadde, K.K.; Al-Duroobi, H.; Phan, D.C.; Jafarzadeh, A.; Moghadam, S.V.; Matta, A.; Kapoor, V. Assessment of Concentration, Recovery, and Normalization of SARS-CoV-2 RNA from Two Wastewater Treatment Plants in Texas and Correlation with COVID-19 Cases in the Community. ACS EST Water 2022, 2, 2060–2069. [Google Scholar] [CrossRef] [PubMed]
- Hassard, F.; Lundy, L.; Singer, A.C.; Grimsley, J.; Di Cesare, M. Innovation in wastewater near-source tracking for rapid identification of COVID-19 in schools. Lancet Microbe 2021, 2, e4–e5. [Google Scholar] [CrossRef] [PubMed]
- Conway, M.J.; Kado, S.; Kooienga, B.K.; Sarette, J.S.; Kirby, M.H.; Marten, A.D.; Ward, A.S.; Abel, J.D.; King, S.; Billette, J.; et al. SARS-CoV-2 wastewater monitoring in rural and small metropolitan communities in Central Michigan. Sci. Total Environ. 2023, 894, 165013. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Valmond, L.; Thomas, J.; Kim, A.; Austin, P.; Foster, M.; Matthews, J.; Kim, P.; Newman, J. Wastewater surveillance in smaller college communities may aid future public health initiatives. PLoS ONE 2022, 17, e270385. [Google Scholar] [CrossRef] [PubMed]
- Peccia, J.; Coleman, C.K.; LaMontagne, C.D.; Miller, M.E.; Kothegal, N.P.; Holcomb, D.A.; Blackwood, A.D.; Clerkin, T.J.; Serre, M.L. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020, 38, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- Dejus, B.; Cacivkins, P.; Gudra, D.; Dejus, S.; Ustinova, M.; Roga, A.; Strods, M.; Kibilds, J.; Boikmanis, G.; Ortlova, K.; et al. Wastewater-based prediction of COVID-19 cases using a random forest algorithm with strain prevalence data: A case study of five municipalities in Latvia. Sci. Total Environ. 2023, 891, 164519. [Google Scholar] [CrossRef]
- Ahmed, W.; Tscharke, B.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Choi, P.; Clarke, L.; Dwyer, J.; Edson, J.; Nguyen, T.M.H.; et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci. Total Environ. 2021, 761, 144216. [Google Scholar] [CrossRef]
- Bibby, K.; Bivins, A.; Wu, Z.; North, D. Making waves: Plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 2021, 202, 117438. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, P.M.; Graber, T.E.; Mercier, E.; Montpetit, D.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T.; et al. Catching a resurgence: Increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021, 770, 145319. [Google Scholar] [CrossRef] [PubMed]
- Krivoňáková, N.; Šoltýsov, A.; Tamáš, M. Mathematical modeling based on RT-qPCR analysis of SARS-CoV-2 in wastewater as a tool for epidemiology. Sci. Rep. 2021, 11, 19456. [Google Scholar] [CrossRef]
- Giacobbo, A.; Rodrigues, M.A.S.; Ferreira, J.Z.; Bernardes, A.M.; de Pinho, M.N. A critical review on SARS-CoV-2 infectivity in water and wastewater. What do we know? Sci. Total Environ. 2021, 774, 145721. [Google Scholar] [CrossRef]
- Kumar, M.; Jiang, G.; Kumar Thakur, A.; Chatterjee, S.; Bhattacharya, T.; Mohapatra, S.; Chaminda, T.; Kumar Tyagi, V.; Vithanage, M.; Bhattacharya, P.; et al. Lead time of early warning by wastewater surveillance for COVID-19: Geographical variations and impacting factors. Chem. Eng. J. 2022, 441, 135936. [Google Scholar] [CrossRef] [PubMed]
- Saawarn, B.; Hait, S. Occurrence, fate and removal of SARS-CoV-2 in wastewater: Current knowledge and future perspectives. J. Environ. Chem. Eng. 2021, 9, 104870. [Google Scholar] [CrossRef] [PubMed]
- Riediker, M.; Briceno-Ayala, L.; Ichihara, G.; Albani, D.; Poffet, D.; Tsai, D.H.; Iff, S.; Monn, C. Higher viral load and infectivity increase risk of aerosol transmission for Delta and Omicron variants of SARS-CoV-2. Swiss Med. Wkly. 2022, 152, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Prasek, S.M.; Pepper, I.L.; Innes, G.K.; Slinski, S.; Betancourt, W.Q.; Foster, A.R.; Yaglom, H.D.; Porter, W.T.; Engelthaler, D.M.; Schmitz, B.W. Variant-specific SARS-CoV-2 shedding rates in wastewater. Sci. Total Environ. 2023, 857, 159165. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, M.; Wei, F.; Huang, S.; Xu, J. COVID’s future: Viral multi-lineage evolution and the dynamics of small epidemic waves without seasonality in COVID-19. J. Biosaf. Biosecur. 2023, 5, 96–99. [Google Scholar] [CrossRef]
- Madewell, Z.J.; Yang, Y.; Longini, I.M.; Halloran, M.E.; Vespignani, A.; Dean, N.E. Rapid review and meta-analysis of serial intervals for SARS-CoV-2 Delta and Omicron variants. BMC Infect. Dis. 2023, 23, 429. [Google Scholar] [CrossRef]
- Ward, T.; Glaser, A.; Overton, C.E.; Carpenter, B.; Gent, N.; Seale, A.C. Replacement dynamics and the pathogenesis of the Alpha, Delta and Omicron variants of SARS-CoV-2. Epidemiol. Infect. 2023, 151, e32. [Google Scholar] [CrossRef] [PubMed]
- Ali, W.; Zhang, H.; Wang, Z.; Chang, C.; Javed, A.; Ali, K.; Du, W.; Niazi, N.K.; Mao, K.; Yang, Z. Occurrence of various viruses and recent evidence of SARS-CoV-2 in wastewater systems. J. Hazard. Mater. 2021, 414, 125439. [Google Scholar] [CrossRef] [PubMed]
- Rose, C.; Parker, A.; Jefferson, B.; Cartmell, E. The characterization of feces and urine: A review of the literature to inform advanced treatment technology. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1827–1879. [Google Scholar] [CrossRef] [PubMed]
- Vo, V.; Tillett, R.L.; Chang, C.L.; Gerrity, D.; Betancourt, W.Q.; Oh, E.C. SARS-CoV-2 variant detection at a university dormitory using wastewater genomic tools. Sci. Total Environ. 2022, 805, 149930. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Phan, N.; Tandukar, S.; Ashoori, R.; Thakali, O.; Mousazadesh, M.; Dehghani, M.H.; Sherchan, S.P. Persistence and occurrence of SARS-CoV-2 in water and wastewater environments: A review of the current literature. Environ. Sci. Pollut. Res. 2022, 29, 85658–85668. [Google Scholar] [CrossRef] [PubMed]
- Rimoldi, S.G.; Stefani, F.; Gigantiello, A.; Polesello, S.; Comandatore, F.; Mileto, D.; Maresca, M.; Longobardi, C.; Mancon, A.; Romeri, F.; et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020, 744, 140911. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bivins, A.; Bertsch, P.M.; Bibby, K.; Choi, P.M.; Farkas, K.; Gyawali, P.; Hamilton, K.A.; Haramoto, E.; Kitajima, M.; et al. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health 2020, 17, 82–93. [Google Scholar] [CrossRef]
- Qasim, S.R. Wastewater Treatment Plants Planning, Design, and Operation, 2nd ed.; Technomic Publishing Company: Boca Raton, FL, USA, 1998. [Google Scholar]
- Heaton, K.W.; Radvan, J.; Cripps, H.; Mountford, R.A.; Braddon, F.E.M.; Hughes, A.O. Defecation frequency and timing, and stool form in the general population: A prospective study. Gut 1992, 33, 818–824. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Bivins, A.; Bibby, K.; Farkas, K.; Gathercole, A.; Haramoto, E.; Gyawali, P.; Korajkic, A.; McMinn, B.R.; et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020, 739, 139960. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Bibby, K.; Haramoto, E.; Hewitt, J.; Huygens, F.; Gyawali, P.; Korajkic, A.; Riddell, S.; Sherchan, S.P.; et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020, 191, 110092. [Google Scholar] [CrossRef]
- Jafferali, M.H.; Khatami, K.; Atasoy, M.; Birgersson, M.; Williams, C.; Cetecioglu, Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2021, 755, 142939. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.R.; Rhodes, E.R.; Brinkman, N.; Wymer, L.; Fout, G.S. New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Appl. Environ. Microbiol. 2009, 75, 2393–2399. [Google Scholar] [CrossRef]
- Dejus, B.; Ozols, R.; Strods, M.; Laicans, J.; Zajakina, A.; Roga, A.; Fridmanis, D.; Juhna, T. Performance Evaluation of Wastewater Concentration Device: Analysis of Recovery Rate for Implementing SARS-CoV-2 Wastewater-Based Epidemiology. Chem. Eng. Trans. 2023, 98, 273–278. [Google Scholar] [CrossRef]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E.; Rose, J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total Environ. 2020, 739, 139076. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors-occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Qiu, Y.; Yu, J.; Pabbaraju, K.; Lee, B.E.; Gao, T.; Ashbolt, N.J.; Hrudey, S.E.; Diggle, M.; Tipples, G.; Maal-Bared, R.; et al. Validating and optimizing the method for molecular detection and quantification of SARS-CoV-2 in wastewater. Sci. Total Environ. 2022, 812, 151434. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, H.D.; Kennedy, L.C.; Hinkle, A.; Whitney, O.N.; Fan, V.B.; Crits-Christoph, A.; Harris-Lovett, S.; Flamholz, A.I.; Al-Shayeb, B.; Liao, L.D.; et al. Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. Water Res. X 2021, 12, 100111. [Google Scholar] [CrossRef] [PubMed]
- Black, J.; Aung, P.; Nolan, M.; Roney, E.; Poon, R.; Hennessy, D.; Crosbie, N.D.; Deere, D.; Jex, A.R.; John, N.; et al. Epidemiological evaluation of sewage surveillance as a tool to detect the presence of COVID-19 cases in a low case load setting. Sci. Total Environ. 2021, 786, 147469. [Google Scholar] [CrossRef]
- Hsu, S.Y.; Bayati, M.B.; Li, C.; Hsieh, H.Y.; Belenchia, A.; Klutts, J.; Zemmer, S.A.; Reynolds, M.; Semkiw, E.; Johnson, H.Y.; et al. Biomarkers selection for population normalization in SARS-CoV-2 wastewater-based epidemiology. Water Res. 2022, 223, 118985. [Google Scholar] [CrossRef]
- Rainey, A.L.; Liang, S.; Bisesi, J.H.; Sabo-Attwood, T.; Maurelli, A.T. A multistate assessment of population normalization factors for wastewater-based epidemiology of COVID-19. PLoS ONE 2023, 18, e0284370. [Google Scholar] [CrossRef] [PubMed]
- Oloye, F.F.; Xie, Y.; Challis, J.K.; Femi-Oloye, O.P.; Brinkmann, M.; McPhedran, K.N.; Jones, P.D.; Servos, M.R.; Giesy, J.P. Understanding common population markers for SARS-CoV-2 RNA normalization in wastewater—A review. Chemosphere 2023, 333, 138682. [Google Scholar] [CrossRef] [PubMed]
- Maal-Bared, R.; Qiu, Y.; Li, Q.; Gao, T.; Hrudey, S.E.; Bhavanam, S.; Ruecker, N.J.; Ellehoj, E.; Lee, B.E.; Pang, X. Does normalization of SARS-CoV-2 concentrations by Pepper Mild Mottle Virus improve correlations and lead time between wastewater surveillance and clinical data in Alberta (Canada) comparing twelve SARS-CoV-2 normalization approaches. Sci. Total Environ. 2022, 856, 158964. [Google Scholar] [CrossRef] [PubMed]
- Wade, M.J.; Lo Jacomo, A.; Armenise, E.; Brown, M.R.; Bunce, J.T.; Cameron, G.J.; Fang, Z.; Farkas, K.; Gilpin, D.F.; Graham, D.W.; et al. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: Lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard. Mater. 2022, 424, 127456. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Kostakis, C.; Gerber, J.P.; Tscharke, B.J.; Irvine, R.J.; White, J.M. Towards finding a population biomarker for wastewater epidemiology studies. Sci. Total Environ. 2014, 487, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Lor, E. Measuring biomarkers in wastewater as a new source of epidemiological information: Current state and future perspectives. Environ. Int. 2017, 99, 131–150. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.M.; Rodríguez-Mozaz, S.; Corominas, L.; Petrovic, M. Wastewater-based epidemiology biomarkers: Past, present and future. TrAC-Trends Anal. Chem. 2018, 105, 453–469. [Google Scholar] [CrossRef]
- Saingam, P.; Li, B.; Quoc, B.N.; Jain, T.; Bryan, A.; Winkler, M.K.H. Wastewater surveillance of SARS-CoV-2 at intra-city level demonstrated high resolution in tracking COVID-19 and calibration using chemical indicators. Sci. Total Environ. 2023, 866, 161467. [Google Scholar] [CrossRef] [PubMed]
- Rico, M.; Andrés-Costa, M.J.; Picó, Y. Estimating population size in wastewater-based epidemiology. Valencia metropolitan area as a case study. J. Hazard. Mater. 2017, 323, 156–165. [Google Scholar] [CrossRef]
- Kasprzyk-Hordern, B.; Sims, N.; Farkas, K.; Jagadeesan, K.; Proctor, K.; Wade, M.J.; Jones, D.L. Wastewater-based epidemiology for comprehensive community health diagnostics in a national surveillance study: Mining biochemical markers in wastewater. J. Hazard. Mater. 2023, 450, 130989. [Google Scholar] [CrossRef]
- Sakarovitch, C.; Schlosser, O.; Courtois, S.; Proust-Lima, C.; Couallier, J.; Pétrau, A.; Litrico, X.; Loret, J.F. Monitoring of SARS-CoV-2 in wastewater: What normalization for improved understanding of epidemic trends? J. Water Health 2022, 20, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Wilder, M.L.; Middleton, F.; Larsen, D.A.; Du, Q.; Fenty, A.; Zeng, T.; Insaf, T.; Kilaru, P.; Collins, M.; Kmush, B.; et al. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X 2021, 11, 100100. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; Crank, K.; Greaves, J.; North, D.; Wu, Z.; Bibby, K. Cross-assembly phage and pepper mild mottle virus as viral water quality monitoring tools—Potential, research gaps, and way forward. Curr. Opin. Environ. Sci. Health 2020, 16, 54–61. [Google Scholar] [CrossRef]
- Ahme, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Van Drecht, G.; Bouwman, A.F.; Harrison, J.; Knoop, J.M. Global nitrogen and phosphate in urban wastewater for the period 1970 to 2050. Glob. Biogeochem. Cycles 2009, 23, 1–19. [Google Scholar] [CrossRef]
- Holm, R.H.; Nagarkar, M.; Yeager, R.A.; Talley, D.; Chaney, A.C.; Rai, J.P.; Mukherjee, A.; Rai, S.N.; Bhatnagar, A.; Smith, T. Surveillance of RNase P, PMMoV, and CrAssphage in wastewater as indicators of human fecal concentration across urban sewer neighborhoods, Kentucky. FEMS Microbes 2022, 3, xtac003. [Google Scholar] [CrossRef] [PubMed]
- Thai, P.K.; O’Brien, J.W.; Banks, A.P.W.; Jiang, G.; Gao, J.; Choi, P.M.; Yuan, Z.; Mueller, J.F. Evaluating the in-sewer stability of three potential population biomarkers for application in wastewater-based epidemiology. Sci. Total Environ. 2019, 671, 248–253. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.W.; Banks, A.P.; Novic, A.J.; Mueller, J.F.; Jiang, G.; Ort, C.; Eaglesham, G.; Yuan, Z.; Thai, P.K. Impact of in-Sewer Degradation of Pharmaceutical and Personal Care Products (PPCPs) Population Markers on a Population Model. Environ. Sci. Technol. 2017, 51, 3816–3823. [Google Scholar] [CrossRef] [PubMed]
- Pandopulos, A.J.; Bade, R.; Tscharke, B.J.; O’Brien, J.W.; Simpson, B.S.; White, J.M.; Gerber, C. Application of catecholamine metabolites as endogenous population biomarkers for wastewater-based epidemiology. Sci. Total Environ. 2021, 763, 142992. [Google Scholar] [CrossRef]
- Li, Y.; Miyani, B.; Zhao, L.; Spooner, M.; Gentry, Z.; Zou, Y.; Rhodes, G.; Li, H.; Kaye, A.; Norton, J.; et al. Surveillance of SARS-CoV-2 in nine neighborhood sewersheds in Detroit Tri-County area, United States: Assessing per capita SARS-CoV-2 estimations and COVID-19 incidence. Sci. Total Environ. 2022, 851, 158350. [Google Scholar] [CrossRef]
- Lastra, A.; Botello, J.; Pinilla, A.; Urrutia, J.I.; Canora, J.; Sánchez, J.; Fernández, P.; Candel, F.J.; Zapatero, A.; Ortega, M.; et al. SARS-CoV-2 detection in wastewater as an early warning indicator for COVID-19 pandemic. Madrid region case study. Environ. Res. 2022, 203, 111852. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Li, J.S.; Jin, M.; Zhen, B.; Kong, Q.X.; Song, N.; Xiao, W.J.; Yin, J.; Wei, W.; Wang, G.J.; et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods 2005, 126, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Lipponen, A.; Hokajärvi, A.M.; Luomala, O.; Sarekoski, A.; Rytkönen, A.; Österlund, P.; Al-Hello, H.; Juutinen, A.; Miettinen, I.T.; et al. Detection and quantification of SARS-CoV-2 RNA in wastewater influent in relation to reported COVID-19 incidence in Finland. Water Res. 2022, 215, 118220. [Google Scholar] [CrossRef] [PubMed]
- Petala, M.; Dafou, D.; Kostoglou, M.; Karapantsios, T.; Kanata, E.; Chatziefstathiou, A.; Sakaveli, F.; Kotoulas, K.; Arsenakis, M.; Roilides, E.; et al. A physicochemical model for rationalizing SARS-CoV-2 concentration in sewage. Case study: The city of Thessaloniki in Greece. Sci. Total Environ. 2021, 755, 142855. [Google Scholar] [CrossRef]
- Champredon, D.; Becker, D.; Peterson, S.W. Emergence and spread of SARS-CoV-2 variants of concern in Canada: A retrospective analysis from clinical and wastewater data. BMC Infect. Dis. 2024, 24, 139. [Google Scholar] [CrossRef] [PubMed]
- Bertels, X.; Demeyer, P.; Van den Bogaert, S.; Boogaerts, T.; van Nuijs, A.L.N.; Delputte, P.; Lahousse, L. Factors influencing SARS-CoV-2 RNA concentrations in wastewater up to the sampling stage: A systematic review. Sci. Total Environ. 2022, 820, 153290. [Google Scholar] [CrossRef] [PubMed]
- Kostoglou, M.; Petala, M.; Karapantsios, T.; Dovas, C.; Roilides, E.; Metallidis, S.; Papa, A.; Stylianidis, E.; Papadopoulos, A.; Papaioannou, N. SARS-CoV-2 adsorption on suspended solids along a sewerage network: Mathematical model formulation, sensitivity analysis, and parametric study. Environ. Sci. Pollut. Res. 2021, 29, 11304–11319. [Google Scholar] [CrossRef]
- Jiang, G.; Liu, Y.; Tang, S.; Kitajima, M.; Haramoto, E.; Arora, S.; Choi, P.M.; Jackson, G.; D’Aoust, P.M.; Delatolla, R.; et al. Moving forward with COVID-19: Future research prospects of wastewater-based epidemiology methodologies and applications. Curr. Opin. Environ. Sci. Health 2023, 33, 100458. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laicans, J.; Dejus, B.; Dejus, S.; Juhna, T. Precision and Accuracy Limits of Wastewater-Based Epidemiology—Lessons Learned from SARS-CoV-2: A Scoping Review. Water 2024, 16, 1220. https://doi.org/10.3390/w16091220
Laicans J, Dejus B, Dejus S, Juhna T. Precision and Accuracy Limits of Wastewater-Based Epidemiology—Lessons Learned from SARS-CoV-2: A Scoping Review. Water. 2024; 16(9):1220. https://doi.org/10.3390/w16091220
Chicago/Turabian StyleLaicans, Juris, Brigita Dejus, Sandis Dejus, and Talis Juhna. 2024. "Precision and Accuracy Limits of Wastewater-Based Epidemiology—Lessons Learned from SARS-CoV-2: A Scoping Review" Water 16, no. 9: 1220. https://doi.org/10.3390/w16091220