Adenovirus Core Proteins: Structure and Function
Abstract
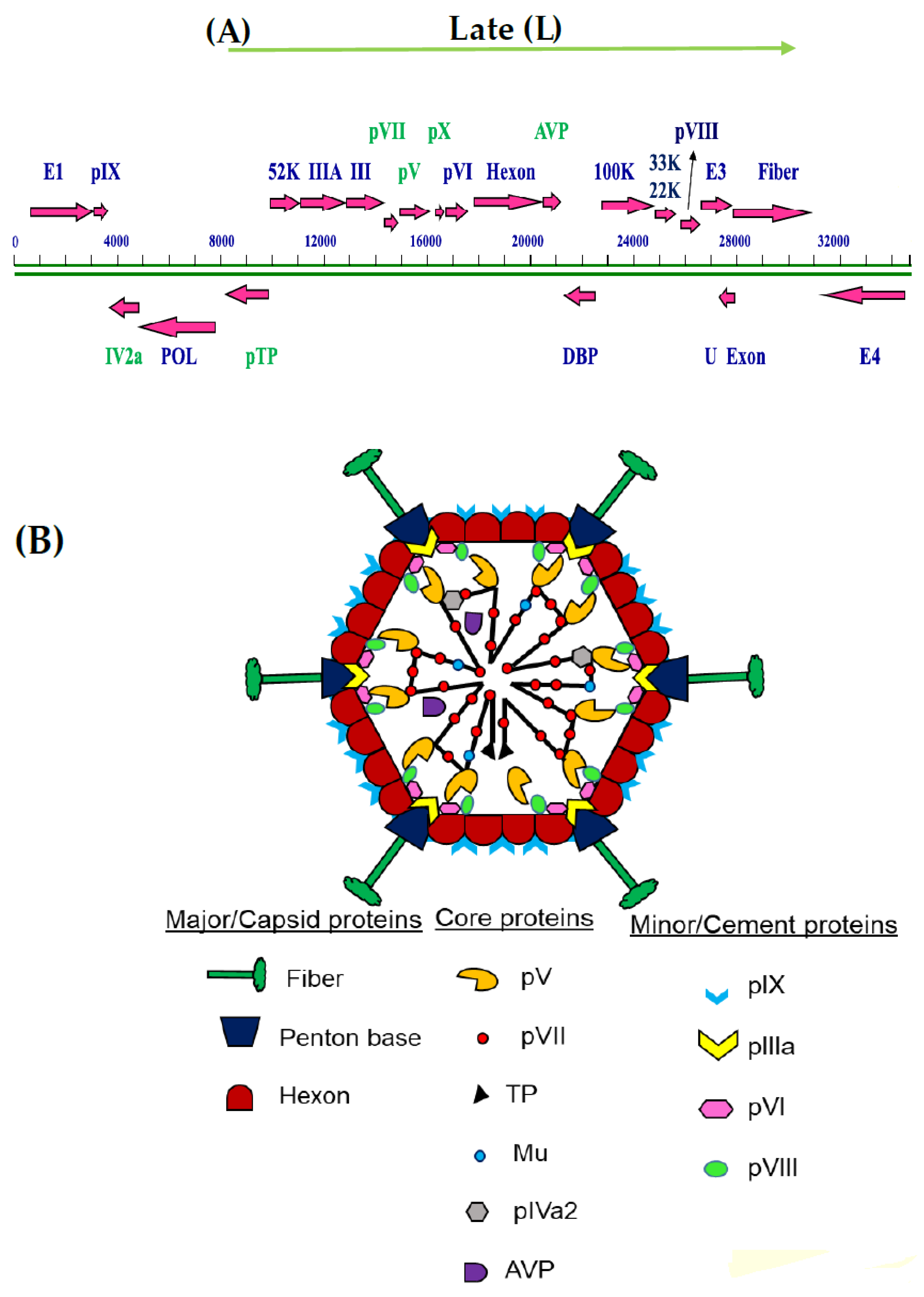
:1. Introduction
2. mAdV Core Proteins
2.1. Protein VII (pVII)
2.1.1. General Characteristics
2.1.2. Functions
2.2. Protein V (pV)
2.2.1. General Characteristics
2.2.2. Functions
2.3. Protein IVa2 (pIVa2)
2.3.1. General Characteristics
2.3.2. Functions
2.4. Adenovirus Protease (Adenain/AVP)
2.5. Protein X (Mu)
2.6. Terminal Protein
3. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Russell, W.C. Adenoviruses: Update on structure and function. J. Gen. Virol. 2009, 90, 1–20. [Google Scholar] [CrossRef]
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef]
- Enders, J.F.; Bell, J.A.; Dingle, J.H.; Francis, T., Jr.; Hilleman, M.R.; Huebner, R.J.; Payne, A.M. Adenoviruses: Group name proposed for new respiratory-tract viruses. Science 1956, 124, 119–120. [Google Scholar] [CrossRef]
- Shenk, T. Adenoviridae: The viruses and their replication. In “Fields Virology”, 4th ed.; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001. [Google Scholar]
- Mese, K.; Bunz, O.; Schellhorn, S.; Volkwein, W.; Jung, D.; Gao, J.; Zhang, W.; Baiker, A.; Ehrhardt, A. Identification of novel human adenovirus candidates using the coxsackievirus and adenovirus receptor for cell entry. Virol. J. 2020, 17, 52. [Google Scholar] [CrossRef] [Green Version]
- Marshall, A.H. Adenoviruses. In “Field’s Virology”, 4th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001; Volume 2, p. 23012326. [Google Scholar]
- Ghebremedhin, B. Human adenovirus: Viral pathogen with increasing importance. Eur. J. Microbiol. Immunol. 2014, 4, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Leen, A.M.; Rooney, C.M. Adenovirus as an emerging pathogen in immunocompromised patients. Br. J. Haematol. 2005, 128, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Tekeleselassie, A.W.; Manoharan, V.K.; Kulanayake, S.; Tikoo, S.K. Adenovirus infections. In Infectious Diseases of Livestock; Coetzer, J.A.W., Thomson, G.R., Maclachlan, N.J., Penrith, M.-L., Eds.; Oxford University Press: Oxford, UK, 2019. [Google Scholar]
- Kajan, G.L.; Affranio, I.; Tothne Bistyak, A.; Kecskemeti, S.; Benko, M. An emerging new fowl adenovirus genotype. Heliyon 2019, 5, e01732. [Google Scholar] [CrossRef] [Green Version]
- Davison, A.J.; Benko, M.; Harrach, B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003, 84 Pt 11, 2895–2908. [Google Scholar] [CrossRef]
- Doszpoly, A.; Wellehan, J.F., Jr.; Childress, A.L.; Tarjan, Z.L.; Kovacs, E.R.; Harrach, B.; Benko, M. Partial characterization of a new adenovirus lineage discovered in testudinoid turtles. Infect. Genet. Evol. 2013, 17, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Fauquet, C.; Briddon, R.; Zerbini, M.; Moriones, E.; Navas-Castillo, J.; King, A.; Adams, M.; Carstens, E.; Lefkowitz, E. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Lefkowitz, E., Adams, M.J., Carstens, E.B., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; p. 351373. [Google Scholar]
- Hoeben, R.C.; Uil, T.G. Adenovirus DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [Green Version]
- Webster, A.; Russell, S.; Talbot, P.; Russell, W.C.; Kemp, G.D. Characterization of the adenovirus proteinase: Substrate specificity. J. Gen. Virol. 1989, 70 Pt 12, 3225–3234. [Google Scholar] [CrossRef]
- Mangel, W.F.; San Martin, C. Structure, function and dynamics in adenovirus maturation. Viruses 2014, 6, 4536–4570. [Google Scholar] [CrossRef] [Green Version]
- Perez-Berna, A.J.; Marion, S.; Chichon, F.J.; Fernandez, J.J.; Winkler, D.C.; Carrascosa, J.L.; Steven, A.C.; Siber, A.; San Martin, C. Distribution of DNA-condensing protein complexes in the adenovirus core. Nucleic Acids Res. 2015, 43, 4274–4283. [Google Scholar] [CrossRef]
- Martin-Gonzalez, N.; Hernando-Perez, M.; Condezo, G.N.; Perez-Illana, M.; Siber, A.; Reguera, D.; Ostapchuk, P.; Hearing, P.; San Martin, C.; de Pablo, P.J. Adenovirus major core protein condenses DNA in clusters and bundles, modulating genome release and capsid internal pressure. Nucleic Acids Res. 2019, 47, 9231–9242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiratsuchi, T.; Rai, U.; Kaneko, I.; Zhang, M.; Iwanaga, S.; Yuda, M.; Tsuji, M. A potent malaria vaccine based on adenovirus with dual modifications at Hexon and pVII. Vaccine 2017, 35, 6990–7000. [Google Scholar] [CrossRef]
- Pfitzner, S.; Hofmann-Sieber, H.; Bosse, J.B.; Franken, L.E.; Grunewald, K.; Dobner, T. Fluorescent protein tagging of adenoviral proteins pV and pIX reveals ‘late virion accumulation compartment’. PLoS Pathog. 2020, 16, e1008588. [Google Scholar] [CrossRef]
- Walkiewicz, M.P.; Morral, N.; Engel, D.A. Accurate single-day titration of adenovirus vectors based on equivalence of protein VII nuclear dots and infectious particles. J. Virol. Methods 2009, 159, 251–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostapchuk, P.; Suomalainen, M.; Zheng, Y.; Boucke, K.; Greber, U.F.; Hearing, P. The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection. PLoS Pathog. 2017, 13, e1006455. [Google Scholar] [CrossRef] [Green Version]
- Reddy, P.S.; Idamakanti, N.; Zakhartchouk, A.N.; Baxi, M.K.; Lee, J.B.; Pyne, C.; Babiuk, L.A.; Tikoo, S.K. Nucleotide sequence, genome organization, and transcription map of bovine adenovirus type 3. J. Virol. 1998, 72, 1394–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, G.; Moria, N.; Williams, M.; Krishnarjuna, B.; Pouton, C.W. Purification and characterization of adenovirus core protein VII: A histone-like protein that is critical for adenovirus core formation. J. Gen. Virol. 2017, 98, 1785–1794. [Google Scholar] [CrossRef]
- Di Francesco, V.; Garnier, J.; Munson, P.J. Improving protein secondary structure prediction with aligned homologous sequences. Protein Sci. 1996, 5, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Mirza, M.A.; Weber, J. Structure of adenovirus chromatin. Biochim. Biophys. Acta 1982, 696, 76–86. [Google Scholar] [CrossRef]
- Callaway, E. Revolutionary cryo-EM is taking over structural biology. Nature 2020, 578, 201. [Google Scholar] [CrossRef] [Green Version]
- Urban, P.L. Quantitative mass spectrometry: An overview. Philos. Trans. A Math. Phys. Eng. Sci. 2016, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avgousti, D.C.; Herrmann, C.; Kulej, K.; Pancholi, N.J.; Sekulic, N.; Petrescu, J.; Molden, R.C.; Blumenthal, D.; Paris, A.J.; Reyes, E.D.; et al. A core viral protein binds host nucleosomes to sequester immune danger signals. Nature 2016, 535, 173–177. [Google Scholar] [CrossRef] [Green Version]
- Weber, J. Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J. Virol. 1976, 17, 462–471. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.M. Adenovirus endopeptidase and its role in virus infection. Curr. Top. Microbiol. Immunol. 1995, 199 Pt 1, 227–235. [Google Scholar]
- Dai, X.; Wu, L.; Sun, R.; Zhou, Z.H. Atomic Structures of Minor Proteins VI and VII in Human Adenovirus. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Ayalew, L.E.; Gaba, A.; Kumar, P.; Tikoo, S.K. Conserved regions of bovine adenovirus-3 pVIII contain functional domains involved in nuclear localization and packaging in mature infectious virions. J. Gen. Virol. 2014, 95, 1743–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inturi, R.; Thaduri, S.; Punga, T. Adenovirus precursor pVII protein stability is regulated by its propeptide sequence. PLoS ONE 2013, 8, e80617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, S.K.; Gaba, A.; Singh, J.; Tikoo, S.K. Bovine adenovirus 3 core protein precursor pVII localizes to mitochondria, and modulates ATP synthesis, mitochondrial Ca2+ and mitochondrial membrane potential. J. Gen. Virol. 2014, 95 Pt 2, 442–452. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.W.R.; Blair, G.E.; Matthews, D.A. Adenovirus core protein VII contains distinct sequences that mediate targeting to the nucleus and nucleolus, and colocalization with human chromosomes. J. Gen. Virol. 2003, 84 Pt 12, 3423–3428. [Google Scholar] [CrossRef]
- Wodrich, H.; Cassany, A.; D’Angelo, M.A.; Guan, T.; Nemerow, G.; Gerace, L. Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J. Virol. 2006, 80, 9608–9618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hindley, C.E.; Lawrence, F.J.; Matthews, D.A. A role for transportin in the nuclear import of adenovirus core proteins and DNA. Traffic 2007, 8, 1313–1322. [Google Scholar] [CrossRef] [Green Version]
- Bremner, K.H.; Scherer, J.; Yi, J.; Vershinin, M.; Gross, S.P.; Vallee, R.B. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 2009, 6, 523–535. [Google Scholar] [CrossRef] [Green Version]
- Suomalainen, M.; Nakano, M.Y.; Keller, S.; Boucke, K.; Stidwill, R.P.; Greber, U.F. Microtubule-Dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999, 144, 657–672. [Google Scholar] [CrossRef] [Green Version]
- Cassany, A.; Ragues, J.; Guan, T.; Begu, D.; Wodrich, H.; Kann, M.; Nemerow, G.R.; Gerace, L. Nuclear import of adenovirus DNA involves direct interaction of hexon with an N-terminal domain of the nucleoporin Nup214. J. Virol. 2015, 89, 1719–1730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlon-Andres, I.; Lagadec, F.; Pied, N.; Rayne, F.; Lafon, M.E.; Kehlenbach, R.H.; Wodrich, H. Nup358 and Transportin 1 Cooperate in Adenoviral Genome Import. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Lischwe, M.A.; Sung, M.T. A histone-like protein from adenovirus chromatin. Nature 1977, 267, 552–554. [Google Scholar] [CrossRef]
- Cheung, P.; Allis, C.D.; Sassone-Corsi, P. Signaling to chromatin through histone modifications. Cell 2000, 103, 263–271. [Google Scholar] [CrossRef] [Green Version]
- Sung, M.T.; Cao, T.M.; Coleman, R.T.; Budelier, K.A. Gene and protein sequences of adenovirus protein VII, a hybrid basic chromosomal protein. Proc. Natl. Acad. Sci. USA 1983, 80, 2902–2906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klune, J.R.; Dhupar, R.; Cardinal, J.; Billiar, T.R.; Tsung, A. HMGB1: Endogenous danger signaling. Mol. Med. 2008, 14, 476–484. [Google Scholar] [CrossRef]
- Bae, J.S. Role of high mobility group box 1 in inflammatory disease: Focus on sepsis. Arch. Pharm. Res. 2012, 35, 1511–1523. [Google Scholar] [CrossRef]
- Karen, K.A.; Hearing, P. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J. Virol. 2011, 85, 4135–4142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiner, S.; Kinkley, S.; Burck, C.; Mund, A.; Wimmer, P.; Schubert, T.; Groitl, P.; Will, H.; Dobner, T. SPOC1-Mediated antiviral host cell response is antagonized early in human adenovirus type 5 infection. PLoS Pathog. 2013, 9, e1003775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernando-Perez, M.; Martin-Gonzalez, N.; Perez-Illana, M.; Suomalainen, M.; Condezo, G.N.; Ostapchuk, P.; Gallardo, J.; Menendez, M.; Greber, U.F.; Hearing, P.; et al. Dynamic competition for hexon binding between core protein VII and lytic protein VI promotes adenovirus maturation and entry. Proc. Natl. Acad. Sci. USA 2020, 117, 13699–13707. [Google Scholar] [CrossRef]
- Johnson, J.S.; Osheim, Y.N.; Xue, Y.; Emanuel, M.R.; Lewis, P.W.; Bankovich, A.; Beyer, A.L.; Engel, D.A. Adenovirus protein VII condenses DNA, represses transcription, and associates with transcriptional activator E1A. J. Virol. 2004, 78, 6459–6468. [Google Scholar] [CrossRef] [Green Version]
- Imperiale, M.J.; Akusjnarvi, G.; Leppard, K.N. Post-Transcriptional control of adenovirus gene expression. Curr. Top. Microbiol. Immunol. 1995, 199 Pt 2, 139–171. [Google Scholar]
- Xue, Y.; Johnson, J.S.; Ornelles, D.A.; Lieberman, J.; Engel, D.A. Adenovirus protein VII functions throughout early phase and interacts with cellular proteins SET and pp32. J. Virol. 2005, 79, 2474–2483. [Google Scholar] [CrossRef] [Green Version]
- Haruki, H.; Okuwaki, M.; Miyagishi, M.; Taira, K.; Nagata, K. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J. Virol. 2006, 80, 794–801. [Google Scholar] [CrossRef] [Green Version]
- Haruki, H.; Gyurcsik, B.; Okuwaki, M.; Nagata, K. Ternary complex formation between DNA-adenovirus core protein VII and TAF-Ibeta/SET, an acidic molecular chaperone. FEBS Lett. 2003, 555, 521–527. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Morral, N.; Engel, D.A. Transcription releases protein VII from adenovirus chromatin. Virology 2007, 369, 411–422. [Google Scholar] [CrossRef] [Green Version]
- Samad, M.A.; Komatsu, T.; Okuwaki, M.; Nagata, K. B23/nucleophosmin is involved in regulation of adenovirus chromatin structure at late infection stages, but not in virus replication and transcription. J. Gen. Virol. 2012, 93 Pt 6, 1328–1338. [Google Scholar] [CrossRef] [Green Version]
- Mun, K.; Punga, T. Cellular Zinc Finger Protein 622 hinders human adenovirus lytic growth and limits binding of the viral pVII protein to virus DNA. J. Virol. 2019, 93, E01628-18. [Google Scholar] [CrossRef] [Green Version]
- Puntener, D.; Engelke, M.F.; Ruzsics, Z.; Strunze, S.; Wilhelm, C.; Greber, U.F. Stepwise loss of fluorescent core protein V from human adenovirus during entry into cells. J. Virol. 2011, 85, 481–496. [Google Scholar] [CrossRef] [Green Version]
- Vayda, M.E.; Rogers, A.E.; Flint, S.J. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 1983, 11, 441–460. [Google Scholar] [CrossRef] [Green Version]
- Burg, J.L.; Schweitzer, J.; Daniell, E. Introduction of superhelical turns into DNA by adenoviral core proteins and chromatin assembly factors. J. Virol. 1983, 46, 749–755. [Google Scholar] [CrossRef] [Green Version]
- Condezo, G.N.; San Martin, C. Localization of adenovirus morphogenesis players, together with visualization of assembly intermediates and failed products, favor a model where assembly and packaging occur concurrently at the periphery of the replication center. PLoS Pathog. 2017, 13, e1006320. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Arcos, R. Interaction of the adenovirus major core protein precursor, pVII, with the viral DNA packaging machinery. Virology 2005, 334, 194–202. [Google Scholar] [CrossRef] [Green Version]
- Perez-Berna, A.J.; Mangel, W.F.; McGrath, W.J.; Graziano, V.; Flint, J.; San Martin, C. Processing of the L1 52/55k Protein by the Adenovirus Protease: A New Substrate and New Insights into Virion Maturation. J. Virol. 2014, 88, 1513–1524. [Google Scholar] [CrossRef] [Green Version]
- Inturi, R.; Mun, K.; Singethan, K.; Schreiner, S.; Punga, T. Human Adenovirus Infection Causes Cellular E3 Ubiquitin Ligase MKRN1 Degradation Involving the Viral Core Protein pVII. J. Virol. 2018, 92, e01154-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, P.K.; Vayda, M.E.; Flint, S.J. Interactions among the three adenovirus core proteins. J. Virol. 1985, 55, 379–386. [Google Scholar] [CrossRef] [Green Version]
- Boudin, M.L.; D’Halluin, J.C.; Cousin, C.; Boulanger, P. Human adenovirus type 2 protein IIIa. II. Maturation and encapsidation. Virology 1980, 101, 144–156. [Google Scholar] [CrossRef]
- Perez-Vargas, J.; Vaughan, R.C.; Houser, C.; Hastie, K.M.; Kao, C.C.; Nemerow, G.R. Isolation and characterization of the DNA and protein binding activities of adenovirus core protein V. J. Virol. 2014, 88, 9287–9296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benevento, M.; Di Palma, S.; Snijder, J.; Moyer, C.; Reddy, V.; Nemerow, G.; Heck, A. Adenovirus composition, proteolysis and disassembly studied by in-depth qualitative and quantitative proteomics. J. Biol. Chem. 2014, 289, 11421–11430. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.S.; Nemerow, G.R. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11715–11720. [Google Scholar] [CrossRef] [Green Version]
- Fedor, M.J.; Daniell, E. Acetylation of histone-like proteins of adenovirus type 5. J. Virol. 1980, 35, 637–643. [Google Scholar] [CrossRef] [Green Version]
- Freudenberger, N.; Meyer, T.; Groitl, P.; Dobner, T.; Schreiner, S. Human Adenovirus Core Protein V Is Targeted by the Host SUMOylation Machinery to Limit Essential Viral Functions. J. Virol. 2018, 92, e01451-17. [Google Scholar]
- Greber, U.F.; Willetts, M.; Webster, P.; Helenius, A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 1993, 75, 477–486. [Google Scholar] [CrossRef] [Green Version]
- Matthews, D.A.; Russell, W.C. Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with nucleoli. J. Gen. Virol. 1998, 1671–1675. [Google Scholar] [CrossRef]
- Matthews, D.A.; Russell, W.C. Adenovirus core protein V interacts with p32—A protein which is associated with both the mitochondria and the nucleus. J. Gen. Virol. 1998, 79 Pt 7, 1677–1685. [Google Scholar] [CrossRef]
- Matthews, D.A. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 2001, 75, 1031–1038. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Tikoo, S.K. Nuclear and Nucleolar localization of bovine adenovirus-3 protein V. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Walton, T.H.; Moen, P.T.; Fox, E.; Bodnar, J.W. Interactions of Minute Virus of Mice and Adenovirus with Host Nucleoli. J. Virol. 1989, 63, 3651–3660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, Y.W.; Evans, V.C.; Heesom, K.J.; Lamond, A.I.; Matthews, D.A. Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol. Cell. Proteom. 2010, 9, 117–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ugai, H.; Borovjagin, A.V.; Le, L.P.; Wang, M.; Curiel, D.T. Thermostability/infectivity defect caused by deletion of the core protein V gene in human adenovirus type 5 is rescued by thermo-selectable mutations in the core protein X precursor. J. Mol. Biol. 2007, 366, 1142–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Tikoo, S.K. Deletion of pV affects integrity of capsid causing defect in the infectivity of bovine adenovirus-3. J. Gen. Virol. 2016, 97, 2657–2667. [Google Scholar] [CrossRef] [PubMed]
- Woldemariam, T.; Wang, W.; Said, A.; Tikoo, S.K. Regions of bovine adenovirus-3 IVa2 involved in nuclear/nucleolar localization and interaction with pV. Virology 2020, 546, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jin, L.; Koh, S.B.; Atanasov, I.; Schein, S.; Wu, L.; Zhou, Z.H. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 2010, 329, 1038–1043. [Google Scholar] [CrossRef] [Green Version]
- Samad, M.A.; Okuwaki, M.; Haruki, H.; Nagata, K. Physical and functional interaction between a nucleolar protein nucleopgosmin/B23 and adenovirus basic core proteins. FEBS Lett. 2007, 581, 3283–3288. [Google Scholar] [CrossRef] [Green Version]
- Kulshreshtha, V.; Tikoo, S.K. Interaction of bovine adenovirus-3 33K protein with other viral proteins. Virology 2008, 381, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Saban, S.D.; Silvestry, M.; Nemerow, G.R.; Stewart, P.L. Visualization of a-helices in a 6-Angstrom resolution cryo-electron microscopy structure of adenovirus allows refinement of capsid protein assignments. J. Virol. 2006, 80, 12049–12059. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Chen, M.; Pettersson, U. A new look at adenovirus splicing. Virology 2014, 456–457, 329–341. [Google Scholar] [CrossRef] [Green Version]
- Jehung, J.P.; Kitamura, T.; Yanagawa-Matsuda, A.; Kuroshima, T.; Towfik, A.; Yasuda, M.; Sano, H.; Kitagawa, Y.; Minowa, K.; Shindoh, M.; et al. Adenovirus infection induces HuR relocalization to facilitate virus replication. Biochem. Biophys. Res. Commun. 2018, 495, 1795–1800. [Google Scholar] [CrossRef]
- Chen, H.; Vinnakota, R.; Flint, S.J. Intragenic activating and repressing elements control transcription from the adenovirus IVa2 initiator. Mol. Cell Biol. 1994, 14, 676–685. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Kiefer, J.; Whalen, D.; Flint, S.J. DNA synthesis-dependent relief of repression of transcription from the adenovirus type 2 IVa(2) promoter by a cellular protein. Virology 2003, 314, 394–402. [Google Scholar] [CrossRef] [Green Version]
- Pardo-Mateos, A.; Young, C.S. A 40 kDa isoform of the type 5 adenovirus IVa2 protein is sufficient for virus viability. Virology 2004, 324, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.C.; Maluf, N.K. Characterization of the non-specific DNA binding properties of the Adenoviral IVa2 protein. Biophys. Chem. 2014, 193–194, 1–8. [Google Scholar] [CrossRef]
- Pettersson, U. Encounters with adenovirus. Upasala J. Med. Sci. 2019, 124, 83–93. [Google Scholar] [CrossRef] [Green Version]
- Winter, N.; D’Halluin, J.C. Regulation of the biosynthesis of subgroup C adenovirus protein IVa2. J. Virol. 1991, 65, 5250–5259. [Google Scholar] [CrossRef] [Green Version]
- Christensen, J.B.; Byrd, S.A.; Walker, A.K.; Strahler, J.R.; Andrews, P.C.; Imperiale, M.J. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J. Virol. 2008, 82, 9086–9093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutz, P.; Puvion-Dutilleul, F.; Lutz, Y.; Kedinger, C. Nucleoplasmic and nucleolar distribution of the adenovirus IVa2 gene product. J. Virol. 1996, 70, 3449–3460. [Google Scholar] [CrossRef] [Green Version]
- Mondesert, G.; Tribouley, C.; Kedinger, C. Identification of a novel downstream binding protein implicated in late-phase-specific activation of the adenovirus major late promotor. Nucleic Acids Res. 1992, 20, 3881–3889. [Google Scholar] [CrossRef] [PubMed]
- Tribouley, C.; Lutz, P.; Staub, A.; Kedinger, C. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J. Virol. 1994, 68, 4450–4457. [Google Scholar] [CrossRef] [Green Version]
- Gustin, K.E.; Lutz, P.; Imperiale, M.M. Interaction of the adenovirus L1 52/55-kilodalton ptrotein with the IVa2 gene product during infection. J. Virol. 1996, 70, 6463–6467. [Google Scholar] [CrossRef] [Green Version]
- Backstrom, E.; Kaufmann, K.B.; Lan, X.; Akusjarvi, G. Adenovirus L4-22K stimulates major late transcription by a mechanism requiring the intergenic late-specific transcription factor-binding site. Virus Res. 2010, 151, 220–228. [Google Scholar] [CrossRef]
- Ali, H.; LeRoy, G.; Bridge, G.; Flint, S.J. The adenovirus L4 33-kilodalton protein binds to intragenic sequences of the major late promoter required for late phase-specific stimulation of transcription. J. Virol. 2007, 81, 1327–1338. [Google Scholar] [CrossRef] [Green Version]
- Lutz, P.; Kedinger, C. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J. Virol. 1996, 70, 1396–1405. [Google Scholar] [CrossRef] [Green Version]
- Pardo-Mateos, A.; Young, C.S. Adenovirus IVa2 protein plays an important role in transcription from the major late promoter in vivo. Virology 2004, 327, 50–59. [Google Scholar] [CrossRef] [Green Version]
- Ostapchuk, P.; Almond, M.; Hearing, P. Characterization of Empty adenovirus particles assembled in the absence of a functional adenovirus IVa2 protein. J. Virol. 2011, 85, 5524–5531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Imperiale, M.J. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 2000, 74, 2687–2693. [Google Scholar] [CrossRef] [Green Version]
- Jansen-Durr, P.; Mondesert, G.; Kedinger, C. Replication-Dependent activation of the adenovirus major late promoter is mediated by the increased binding of a transcription factor to sequences in the first intron. J. Virol. 1989, 63, 5124–5132. [Google Scholar] [CrossRef] [Green Version]
- Schmid, S.I.; Hearing, P. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J. Virol. 1997, 71, 3375–3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, J.B.; Ewing, S.G.; Imperiale, M.J. Identification and characterization of a DNA binding domain on the adenovirus IVa2 protein. Virology 2012, 433, 124–130. [Google Scholar] [CrossRef]
- Ewing, S.G.; Byrd, S.A.; Christensen, J.B.; Tyler, R.E.; Imperiale, M.J. Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins. J. Virol. 2007, 81, 12450–12457. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Orozco, D.; Hearing, P. The adenovirus L4-22K protein is multifunctional and is an integral component of crucial aspects of infection. J. Virol. 2012, 86, 10474–10483. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.-C.; Maluf, N.K. Cooperative heteroassembly of the adenoviral L4-22K and IVa2 proteins on to the viral packaging sequence DNA. Biochemistry 2012, 51, 1357–1368. [Google Scholar] [CrossRef]
- Tyler, R.E.; Ewing, S.G.; Imperiale, M.J. Formation of a multiple protein complex on the adenovirus packaging sequence by the IVa2 protein. J. Virol. 2007, 81, 3447–3454. [Google Scholar] [CrossRef] [Green Version]
- Ostapchuk, P.; Hearing, P. Adenovirus IVa2 protein binds ATP. J. Virol. 2008, 82, 10290–10294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leipe, D.D.; Koonin, E.V.; Aravind, L. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003, 333, 781–815. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Iyer, L.M.; Aravind, L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007, 3, 48–65. [Google Scholar] [PubMed]
- Koonin, E.V. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 1993, 229, 1165–1174. [Google Scholar] [CrossRef]
- Ahi, Y.S.; Vemula, S.V.; Hassan, A.O.; Costakes, G.; Stauffacher, C.; Mittal, S.K. Adenoviral L4 33K forms ring-like oligomers and stimulates ATPase activity of IVa2: Implications in viral genome packaging. Front. Microbiol. 2015, 6, 318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Imperiale, M.J. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 2003, 77, 3586–3594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parent, K.N.; Schrad, J.R.; Cingolani, G. Breaking Symmetry in Viral Icosahedral Capsids as Seen through the Lenses of X-ray Crystallography and Cryo-Electron Microscopy. Viruses 2018, 10, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahi Chelikani, V.; Ranjan, T.; Kondabagil, K. Revisiting the genome packaging in viruses with lessons from the “Giants”. Virology 2014, 466–467, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Ahi, Y.S.; Hassan, A.O.; Vemula, S.V.; Li, K.P.; Jiang, W.; Zhang, G.J.; Mittal, S.K. Adenoviral E4 34K protein interacts with virus packaging components and may serve as the putative portal. Sci. Rep. UK 2017, 7, 7582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostapchuk, P.; Anderson, M.E.; Chandrasekhar, S.; Hearing, P. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 2006, 80, 6973–6981. [Google Scholar] [CrossRef] [Green Version]
- Ahi, Y.S.; Vemula, S.V.; Mittal, S.K. Adenoviral E2 IVa2 protein interacts with L4 33K protein and E2 DNA-binding protein. J. Gen. Virol. 2013, 94 Pt 6, 1325–1334. [Google Scholar] [CrossRef]
- Honkavuori, K.S.; Pollard, B.D.; Rodriguez, M.S.; Hay, R.T.; Kemp, G.D. Dual role of the adenovirus pVI C terminus as a nuclear localization signal and activator of the viral protease. J. Gen. Virol. 2004, 85 Pt 11, 3367–3376. [Google Scholar] [CrossRef]
- Ruzindana-Umunyana, A.; Sircar, S.; Weber, J.M. The effect of mutant peptide cofactors on adenovirus protease activity and virus infection. Virology 2000, 270, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.Z.; McGrath, W.J.; Sweet, R.M.; Mangel, W.F. Crystal structure of the human adenovirus proteinase with its 11 amino acid cofactor. EMBO J. 1996, 15, 1778–1783. [Google Scholar] [CrossRef] [PubMed]
- Diouri, M.; Keyvani-Amineh, H.; Geoghegan, K.F.; Weber, J.M. Cleavage efficiency by adenovirus protease is site-dependent. J. Biol. Chem. 1996, 271, 32511–32514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, P.K.; Flint, S.J. Adenovirus type 2 endopeptidase: An unusual phosphoprotein enzyme matured by autocatalysis. Proc. Natl. Acad. Sci. USA 1987, 84, 714–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, M.T.; McGrath, W.J.; Toledo, D.L.; Mangel, W.F. Different modes of inhibition of human adenovirus proteinase, probably a cysteine proteinase, by bovine pancreatic trypsin inhibitor. FEBS Lett. 1996, 388, 233–237. [Google Scholar] [CrossRef] [Green Version]
- Mangel, W.F.; McGrath, W.J.; Toledo, D.L.; Anderson, C.W. Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature 1993, 361, 274–275. [Google Scholar] [CrossRef] [PubMed]
- Graziano, V.; Luo, G.B.; Blainey, P.C.; Perez-Berna, A.J.; Mcgrath, W.J.; Flint, S.J.; Martin, C.S.; Xie, X.S.; Mangel, W.F. Regulation of a Viral Proteinase by a Peptide and DNA in One-dimensional Space II. adenovirus proteinase is activated in an unusual one-dimensional biochemical reaction. J. Biol. Chem. 2013, 288, 2068–2080. [Google Scholar] [CrossRef] [Green Version]
- Blainey, P.C.; Graziano, V.; Perez-Berna, A.J.; McGrath, W.J.; Flint, S.J.; Martin, C.S.; Xie, X.S.; Mangel, W.F. Regulation of a Viral Proteinase by a Peptide and DNA in One-dimensional Space IV. VIRAL proteinase slides along DNA to locate and process its substrates. J. Biol. Chem. 2013, 288, 2092–2102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangel, W.F.; McGrath, W.J.; Xiong, K.; Graziano, V.; Blainey, P.C. Molecular sled is an eleven-amino acid vehicle facilitating biochemical interactions via sliding components along DNA. Nat. Commun. 2016, 7, 10202. [Google Scholar] [CrossRef] [Green Version]
- Greber, U.F.; Webster, P.; Helenius, A. The role of the adenovirus protease on virus entry into cells. EMBO J. 1996, 15, 1766–1777. [Google Scholar] [CrossRef] [Green Version]
- Makadiya, N.; Gaba, A.; Tikoo, S.K. Cleavage of bovine adenovirus type 3 non-structural 100K protein by protease is required for nuclear localization in infected cells but is not essential for virus replication. J. Gen. Virol. 2015, 96, 2749–2763. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.T.; McBride, K.M.; Baniecki, M.L.; Reich, N.C.; Marriott, G.; Mangel, W.F. Actin can act as a cofactor for a viral proteinase in the cleavage of the cytoskeleton. J. Biol. Chem. 2002, 277, 46298–46303. [Google Scholar] [CrossRef] [Green Version]
- Baniecki, M.L.; McGrath, W.J.; Mangel, W.F. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space: III. Atomic resolution structure of the nascent form of the adenovirus proteinase. J. Biol. Chem. 2013, 288, 2081–2091. [Google Scholar] [CrossRef] [Green Version]
- McGrath, W.J.; Ding, J.; Sweet, R.M.; Mangel, W.F. Crystallographic structure at 1.6-Å resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: Insights on a new fold. Biochem. Biophys. Acta 2003, 1648, 1–11. [Google Scholar] [CrossRef]
- McGrath, W.J.; Graziano, V.; Mangel, W.F. First generation inhibitors of the adenovirus proteinase. FEBS Lett. 2013, 587, 2332–2339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, C.W.; Young, M.E.; Flint, S.J. Characterization of the adenovirus 2 virion protein, mu. Virology 1989, 172, 506–512. [Google Scholar] [CrossRef]
- Lee, T.W.R.; Lawrence, F.J.; Dauksaite, V.; Akusjarvi, G.; Blair, G.E.; Matthews, D.A. Precursor of human adenovirus core polypeptide Mu targets the nucleolus and modulates the expression of E2 proteins. J. Gen. Virol. 2004, 85 Pt 1, 185–196. [Google Scholar] [CrossRef]
- Freimuth, P.; Anderson, C.W. Human Adenovirus Serotype-12 Virion Precursors Pmu and Pvi Are Cleaved at Amino-Terminal and Carboxy-Terminal Sites That Conform to the Adenovirus-2 Endoproteinase Cleavage Consensus Sequence. Virology 1993, 193, 348–355. [Google Scholar] [CrossRef]
- Weber, J.M.; Anderson, C.W. Identification of the gene coding for the precursor of adenovirus core protein X. J. Virol. 1988, 62, 1741–1745. [Google Scholar] [CrossRef] [Green Version]
- Murray, K.D.; Etheridge, C.J.; Shah, S.I.; Matthews, D.A.; Russell, W.; Gurling, H.M.D.; Miller, A.D. Enhanced cationic liposome-mediated transfection using the DNA-binding peptide mu (mu) from the adenovirus core. Gene Ther. 2001, 8, 453–460. [Google Scholar] [CrossRef] [Green Version]
- Rekosh, D.M.K.; Russell, W.C.; Bellet, A.J.D.; Robinson, A.J. Identification of a Protein Linked to Ends of Adenovirus DNA. Cell 1977, 11, 283–295. [Google Scholar] [CrossRef]
- Pronk, R.; Stuiver, M.H.; Vandervliet, P.C. Adenovirus DNA-Replication—The Function of the Covalently Bound Terminal Protein. Chromosoma 1992, 102, S39–S45. [Google Scholar] [CrossRef] [PubMed]
- Coenjaerts, F.E.; van Oosterhout, J.A.; van der Vliet, P.C. The Oct-1 POU domain stimulates adenovirus DNA replication by a direct interaction between the viral precursor terminal protein-DNA polymerase complex and the POU homeodomain. EMBO J. 1994, 13, 5401–5409. [Google Scholar] [CrossRef] [PubMed]
- Guggenheimer, R.A.; Nagata, K.; Kenny, M.; Hurwitz, J. Protein-Primed replication of plasmids containing the terminus of the adenovirus genome. II. Purification and characterization of a host protein required for the replication of DNA templates devoid of the terminal protein. J. Biol. Chem. 1984, 259, 7815–7825. [Google Scholar] [CrossRef]
| Mastadenovirus Protein | Functions |
|---|---|
| VII | Nuclear transport of the viral genome [37,42] Acts as the cellular histone [26,29,43,44] Functionally mimics protamine [45] Prevents induction of innate immune response [29] Prevents recognition of AdV DNA-by-DNA damage sensor MRN [48] Counteracts SPOC-1-mediated antiviral response [49] Is involved directly/indirectly in the endosomal escape of uncoated virions and pVI cleavage by protease [22,50] Facilitates early gene transcription [51,52,55,56] Prevents deposition of the cellular histones on newly replicated AdV genome [55] Remodels viral chromatin at late times post-infection [57] Hinders viral replication at late times post-infection [58] Condenses AdV genomic DNA [29,51,59,60,61] N-terminus of pVII may act as an anchor to assemble capsomers around the condensed AdV genome [32] Facilitates efficient viral production by inhibiting apoptosis [35] or inhibitory effect of the cellular protein [65] |
| V | Produces of stable progeny virions [81] Interacts with the viral DNA and other core proteins [66,68,75,82] ii) Interacts with other viral capsid proteins [17,66,67,68,70,75] May be required for expression of late gene expression [80,81] May be involved in AdV genome condensation [24,59,68,83] May be involved in mRNA transcription [66], DNA encapsidation [82] and/or virus assembly [84] Other roles in virus replication [67,85,86] |
| IVa2 | Activates AdV major late promoter [97,98,99,100,101,102] Packages adenovirus DNA [109,110,111,112] Acts as DNA packaging ATPase [104,113,114,115,116,126] and involved in the insertion of the viral DNA in empty capsids [118] |
| Protease | Is essential for virus maturation and production of infectious progeny virion [31] Is esential for the proper release of the incoming uncoated virion to the cytoplasm [134] Cleaves precursor adenoviral proteins IIIA, VI, VIII, Mu/X, TP and 52K/55K in virion [16,64] Cleaves 100K in the cytoplasm of transfected cells [135] |
| Mu/X | Condenses AdV genome [27] Alters accumulation of E2 proteins [141] Is involved in increasing DNA transfection efficiency [143] |
| Terminal Protein | Acts as a primer for DNA replication [146,147] Protects AdV DNA from nuclease activity [146] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulanayake, S.; Tikoo, S.K. Adenovirus Core Proteins: Structure and Function. Viruses 2021, 13, 388. https://doi.org/10.3390/v13030388
Kulanayake S, Tikoo SK. Adenovirus Core Proteins: Structure and Function. Viruses. 2021; 13(3):388. https://doi.org/10.3390/v13030388
Chicago/Turabian StyleKulanayake, Shermila, and Suresh K. Tikoo. 2021. "Adenovirus Core Proteins: Structure and Function" Viruses 13, no. 3: 388. https://doi.org/10.3390/v13030388
APA StyleKulanayake, S., & Tikoo, S. K. (2021). Adenovirus Core Proteins: Structure and Function. Viruses, 13(3), 388. https://doi.org/10.3390/v13030388






