Comprehensive Analysis of Jumonji Domain C Family from Citrus grandis and Expression Profilings in the Exocarps of “Huajuhong” (Citrus grandis “Tomentosa”) during Various Development Stages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of JMJC Genes from Citrus grandis
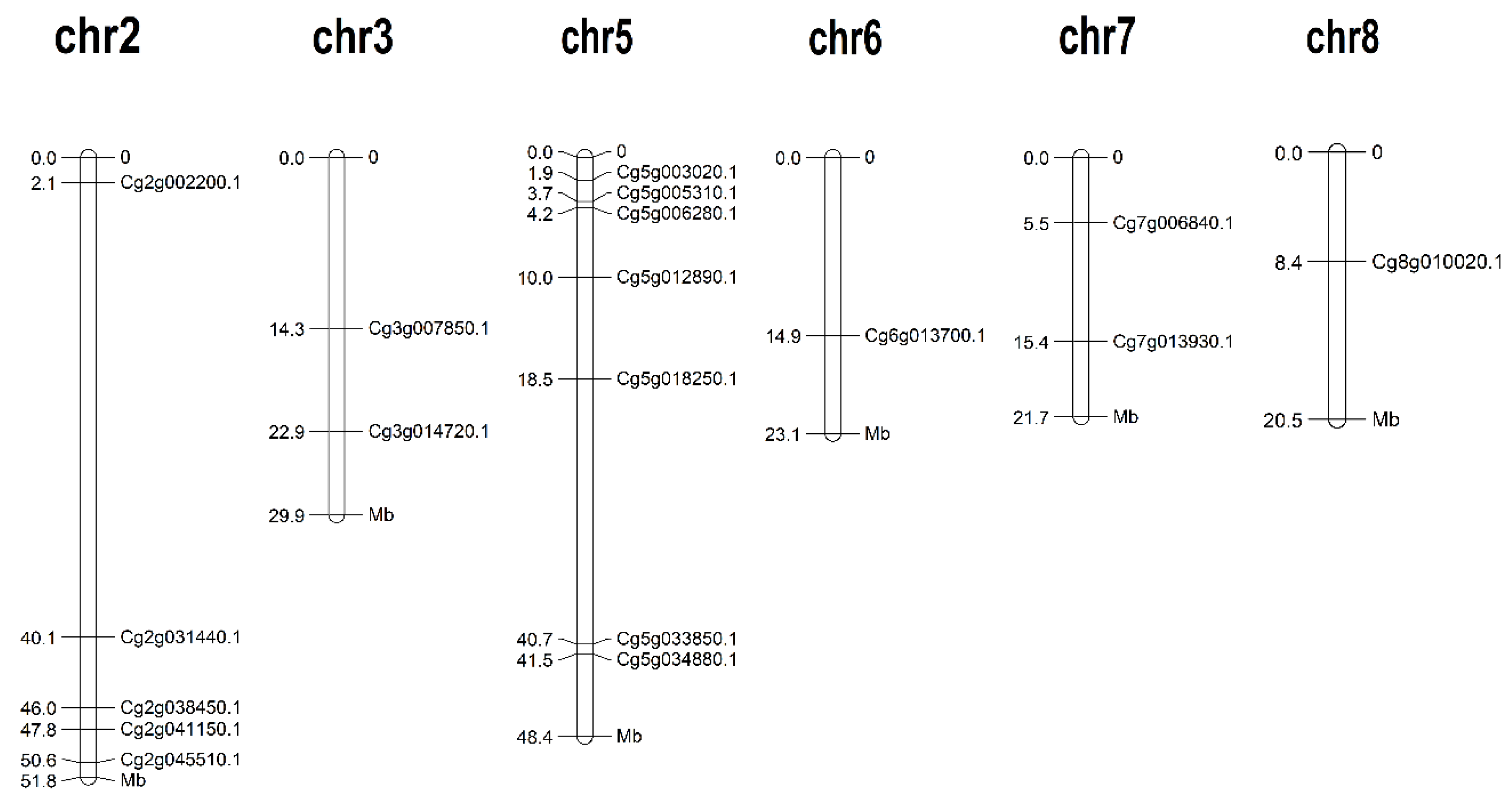
2.2. Chromosomal Localization of CgJMJC Genes
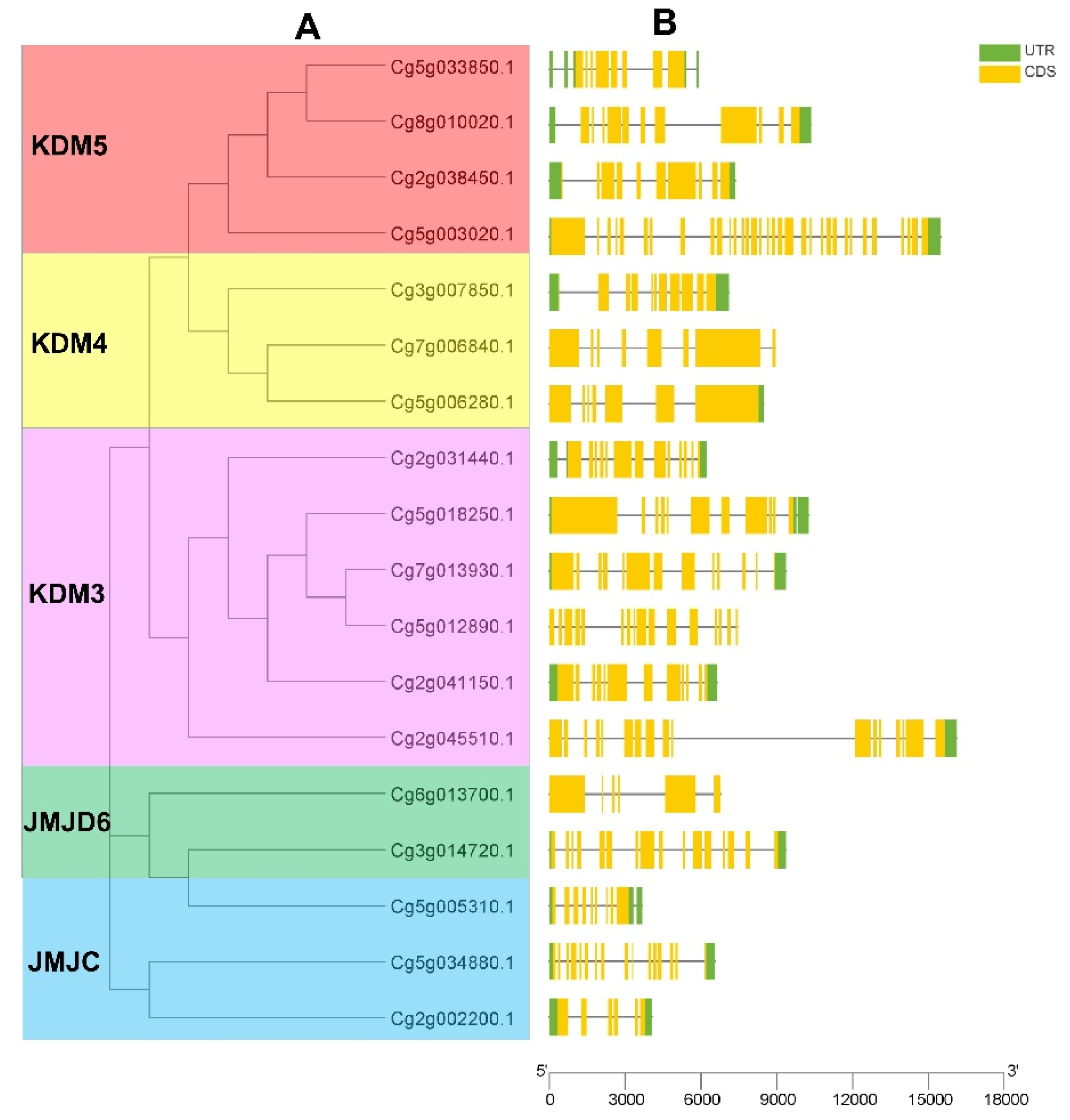
2.3. Phylogenetic Analysis of JMJC Genes from Citrus grandis and Arabidopsis
2.4. Gene Structure Analysis
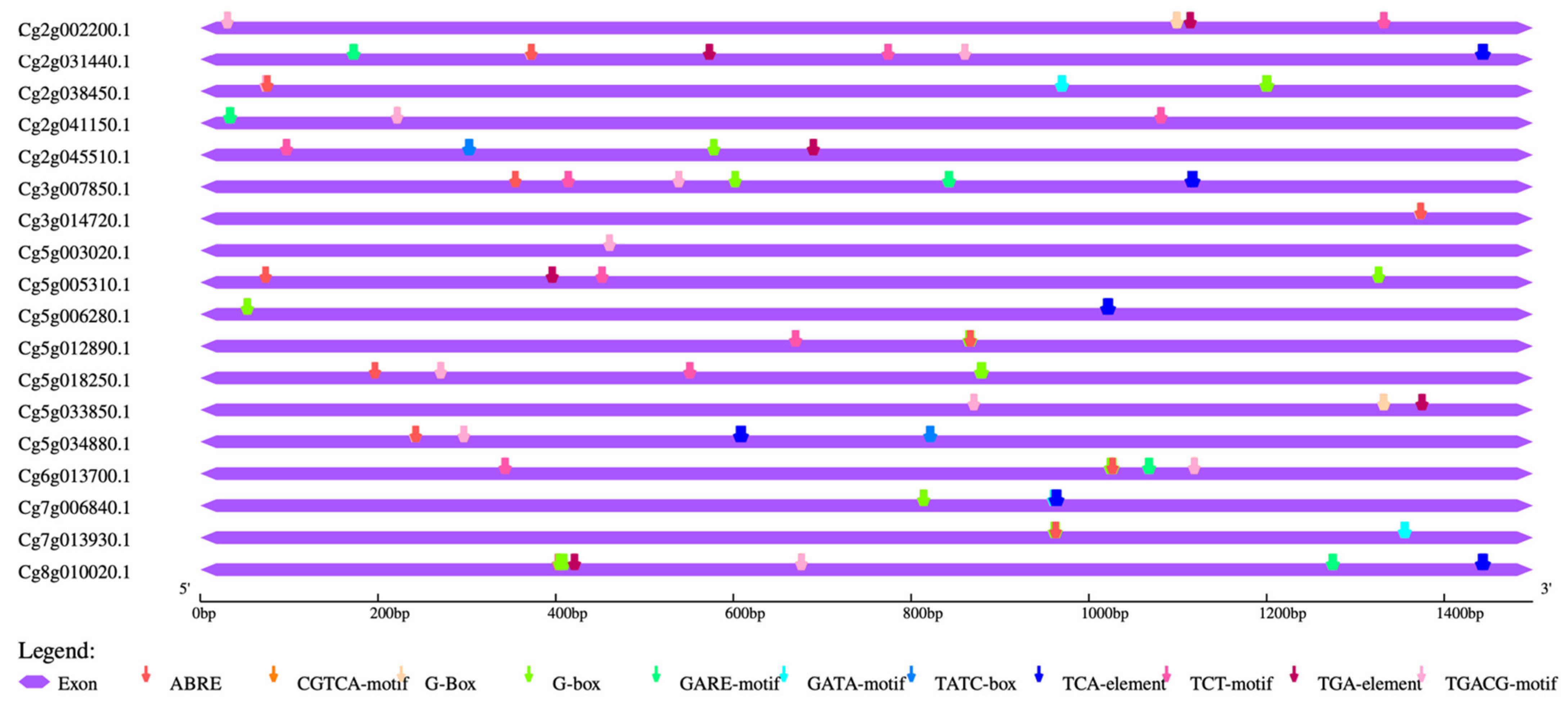
2.5. Cis-Element Analysis and Heat Map Construction
2.6. Plant Material and Treatments
2.7. RNA Isolation and RT-PCR Analysis
3. Results
3.1. Identification and Chromosomal Localization of JMJC Genes from Citrus grandis
3.2. Phylogenetic Analysis of JMJC Genes in Citrus grandis, Citrus sinensis, and Arabidopsis
3.3. Gene Structure Analysis
3.4. Cis-Element Analysis
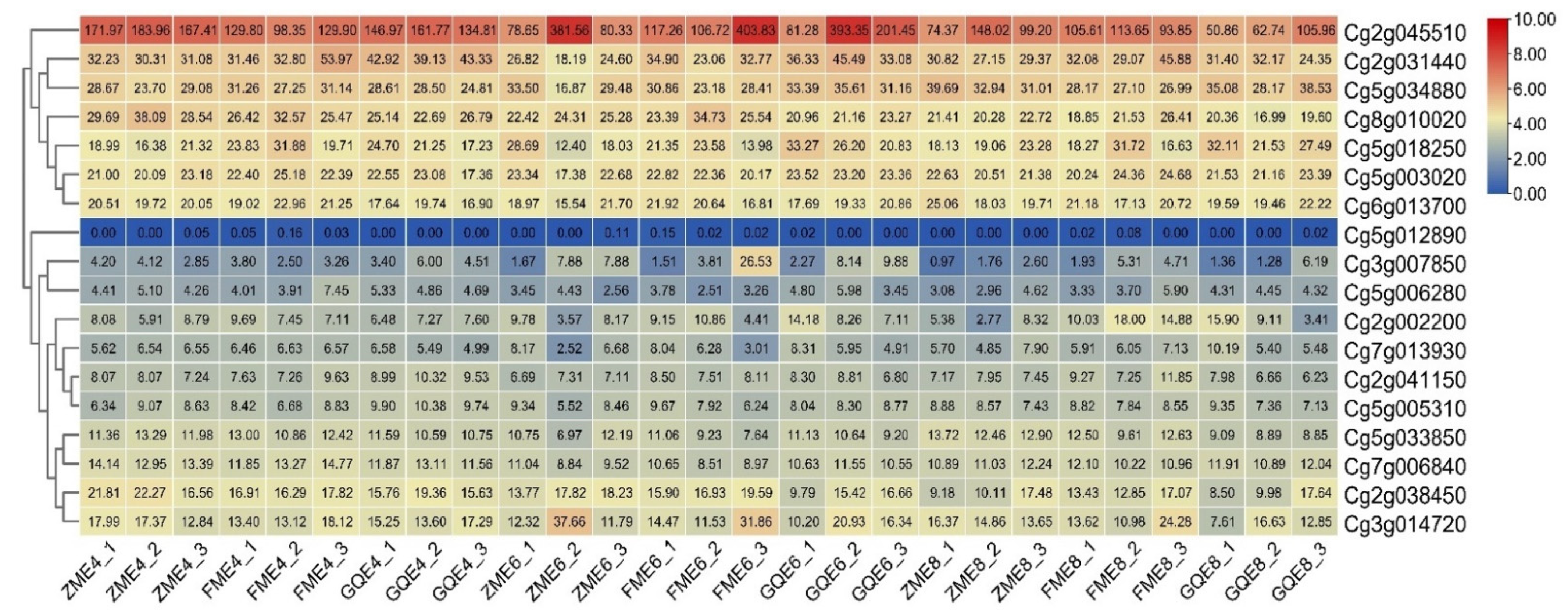
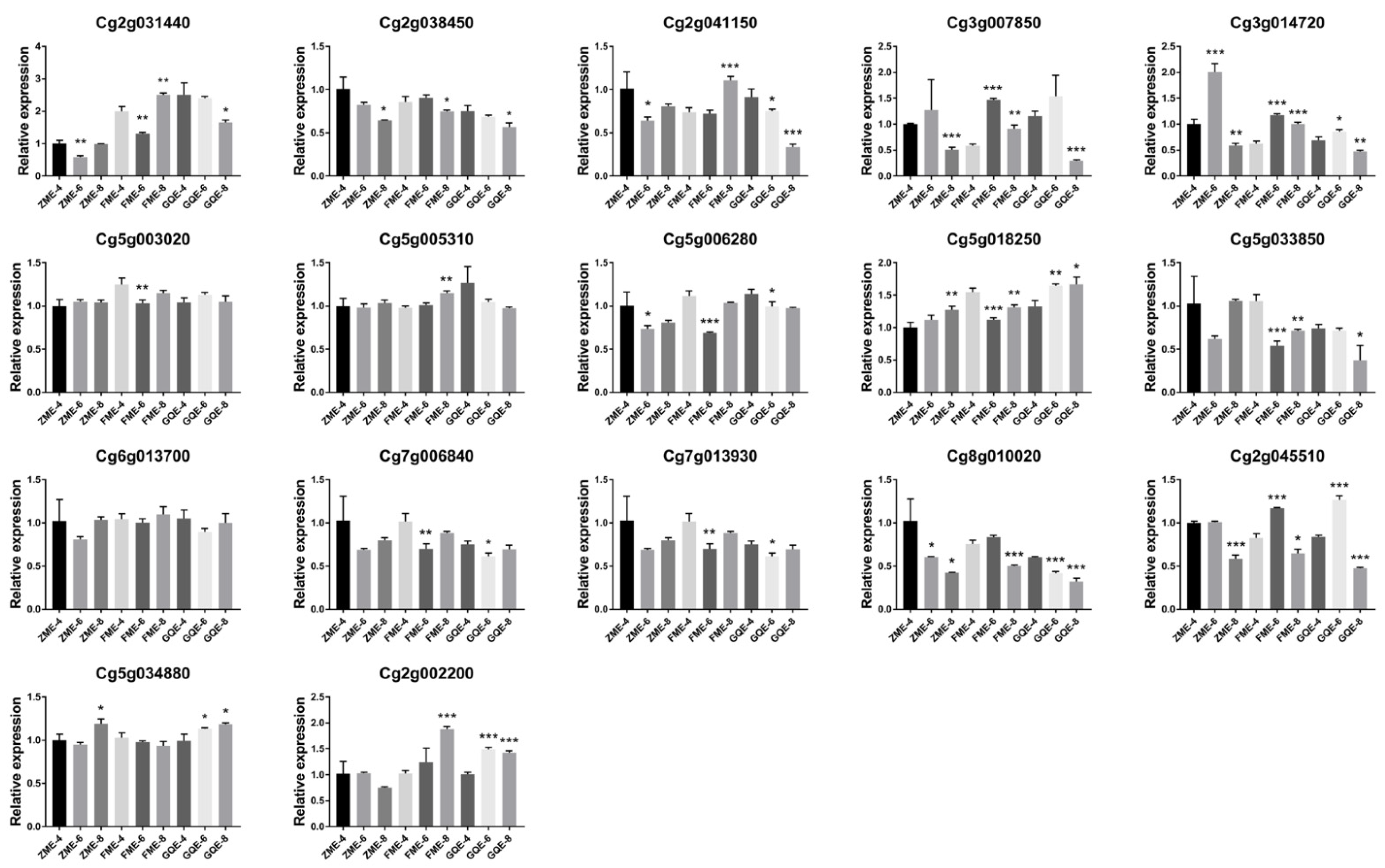
3.5. Expression Patterns in the Exocarps of “Huajuhong” (Citrus grandis “Tomentosa”) during Various Development Stages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Matouk, C.C.; Marsden, P.A. Epigenetic regulation of vascular endothelial gene expression. Circ. Res. 2008, 102, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Richmond, T.J. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998, 8, 140. [Google Scholar] [CrossRef]
- Zhang, X. The epigenetic landscape of plants. Science 2008, 320, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Kanchanaketu, T.; Sangduen, N.; Toojinda, T.; Hongtrakul, V. Genetic diversity analysis of Jatropha curcas L. (Euphorbiaceae) based on methylation-sensitive amplification polymorphism. Genet. Mol. Res. 2012, 11, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Peraza-Echeverria, S.; Herrera-Valencia, V.A.; Kay, A.J. Detection of DNA methylation changes in micropropagated banana plants using methylation-sensitive amplification polymorphism (MSAP). Plant Sci. 2001, 161, 359–367. [Google Scholar] [CrossRef]
- Mastan, S.G.; Rathore, M.S.; Bhatt, V.D.; Yadav, P.; Chikara, J. Assessment of changes in DNA methylation by methylation-sensitive amplification polymorphism in Jatropha curcas L. subjected to salinity stress. Gene 2012, 508, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Hung, F.-Y.; Yang, S.; Liu, X.; Wu, K. Histone Lysine Demethylases and Their Functions in Plants. Plant Mol. Biol. Rep. 2013, 32, 558–565. [Google Scholar] [CrossRef]
- Jiang, D.; Yang, W.; He, Y.; Amasino, R.M. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 2007, 19, 2975–2987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Hu, Y.; Zhou, D.X. Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 2011, 1809, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Zang, J.; Whetstine, J.; Hong, X.; Davrazou, F.; Kutateladze, T.G.; Simpson, M.; Mao, Q.; Pan, C.H.; Dai, S.; et al. Structural insights into histone demethylation by JMJD2 family members. Cell 2006, 125, 691–702. [Google Scholar] [CrossRef] [Green Version]
- Trewick, S.C.; McLaughlin, P.J.; Allshire, R.C. Methylation: Lost in hydroxylation? EMBO Rep. 2005, 6, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Whetstine, J.R.; Nottke, A.; Lan, F.; Huarte, M.; Smolikov, S.; Chen, Z.; Spooner, E.; Li, E.; Zhang, G.; Colaiacovo, M.; et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006, 125, 467–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noh, B.; Lee, S.H.; Kim, H.J.; Yi, G.; Shin, E.A.; Lee, M.; Jung, K.J.; Doyle, M.R.; Amasino, R.M.; Noh, Y.S. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 2004, 16, 2601–2613. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Mo, H.; Fan, D.; Cao, Y.; Cui, S.; Ma, L. Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in Arabidopsis. Plant Cell Rep. 2012, 31, 1297–1308. [Google Scholar] [CrossRef]
- Zheng, S.; Hu, H.; Ren, H.; Yang, Z.; Qiu, Q.; Qi, W.; Liu, X.; Chen, X.; Cui, X.; Li, S.; et al. The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat. Commun. 2019, 10, 1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Zhang, S.; Zhou, B.; Luo, X.; Zhou, X.F.; Cai, B.; Jin, Y.H.; Niu, D.; Lin, J.; Cao, X.; et al. The Histone H3K4 Demethylase JMJ16 Represses Leaf Senescence in Arabidopsis. Plant Cell 2019, 31, 430–443. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.N.; Ryu, J.Y.; Jeong, Y.M.; Park, J.; Song, J.J.; Amasino, R.M.; Noh, B.; Noh, Y.S. Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 2012, 22, 736–748. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Liu, H.; Cheng, Z.J.; Su, Y.H.; Han, H.N.; Zhang, Y.; Zhang, X.S. DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 2011, 7, e1002243. [Google Scholar] [CrossRef] [Green Version]
- Lu, F.; Li, G.; Cui, X.; Liu, C.; Wang, X.J.; Cao, X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 2008, 50, 886–896. [Google Scholar] [CrossRef]
- Han, Y.; Li, X.; Cheng, L.; Liu, Y.; Wang, H.; Ke, D.; Yuan, H.; Zhang, L.; Wang, L. Genome-Wide Analysis of Soybean JmjC Domain-Containing Proteins Suggests Evolutionary Conservation Following Whole-Genome Duplication. Front. Plant Sci. 2016, 7, 1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Y.; Chen, C.; Jiang, L.; Zhang, J.; Ren, Q. Genome-wide identification, classification and expression analysis of the JmjC domain-containing histone demethylase gene family in maize. BMC Genom. 2019, 20, 256. [Google Scholar] [CrossRef]
- Xu, J.; Xu, H.; Liu, Y.; Wang, X.; Xu, Q.; Deng, X. Genome-wide identification of sweet orange (Citrus sinensis) histone modification gene families and their expression analysis during the fruit development and fruit-blue mold infection process. Front. Plant Sci. 2015, 6, 607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Wang, X.; Qiao, K.; Fan, S.; Ma, Q. Genome-wide analysis of JMJ-C histone demethylase family involved in salt-tolerance in Gossypium hirsutum L. Plant Physiol. Biochem. 2021, 158, 420–433. [Google Scholar] [CrossRef]
- Fan, R.; Zhu, C.; Qiu, D.; Zeng, J. Comparison of the bioactive chemical components and antioxidant activities in three tissues of six varieties of Citrus grandis ‘Tomentosa’ fruits. Int. J. Food Prop. 2019, 22, 1848–1862. [Google Scholar] [CrossRef] [Green Version]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.H.; Dhawan, P.; Nath, P. A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Mol. Biol. Rep. 2012, 18, 109–115. [Google Scholar] [CrossRef]
- Li, B.; Fan, R.; Yang, Q.; Hu, C.; Sheng, O.; Deng, G.; Dong, T.; Li, C.; Peng, X.; Bi, F.; et al. Genome-Wide Identification and Characterization of the NAC Transcription Factor Family in Musa Acuminata and Expression Analysis during Fruit Ripening. Int. J. Mol. Sci. 2020, 21, 634. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, T.; Yamazaki, Y.; Katoh-Fukui, Y.; Tsuchiya, R.; Kondo, S.; Motoyama, J.; Higashinakagawa, T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995, 9, 1211–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellagatti, A.; Boultwood, J. Splicing factor gene mutations in the myelodysplastic syndromes: Impact on disease phenotype and therapeutic applications. Adv. Biol. Regul. 2017, 63, 59–70. [Google Scholar] [CrossRef]
- Deeba, F.; Pandey, A.K.; Ranjan, S.; Mishra, A.; Singh, R.; Sharma, Y.K.; Shirke, P.A.; Pandey, V. Physiological and proteomic responses of cotton (Gossypium herbaceum L.) to drought stress. Plant Physiol. Biochem. 2012, 53, 6–18. [Google Scholar] [CrossRef]
- Guo, J.; Wang, S.; Yu, X.; Dong, R.; Li, Y.; Mei, X.; Shen, Y. Polyamines Regulate Strawberry Fruit Ripening by Abscisic Acid, Auxin, and Ethylene. Plant Physiol. 2018, 177, 339–351. [Google Scholar] [CrossRef] [Green Version]
- Li, P.L.; Liu, M.H.; Hu, J.H.; Su, W.W. Systematic chemical profiling of Citrus grandis ‘Tomentosa’ by ultra-fast liquid chromatography/diode-array detector/quadrupole time-of-flight tandem mass spectrometry. J. Pharm. Biomed. Anal. 2014, 90, 167–179. [Google Scholar] [CrossRef]
- Yu, X.; Liu, Q.; Xie, Z.; Lam, S.; Xu, X. Chromatographic Fingerprint Analysis of Exocarpium Citri Grandis by High-Performance Liquid Chromatography Coupled with Diode-Array Detector. Food Anal. Methods 2014, 8, 1868–1875. [Google Scholar] [CrossRef]
- Akimoto, K.; Katakami, H.; Kim, H.J.; Ogawa, E.; Sano, C.M.; Wada, Y.; Sano, H. Epigenetic inheritance in rice plants. Ann. Bot. 2007, 100, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Conde, E.S.N.; Audonnet, L.; Servet, C.; Wei, W.; Zhou, D.X. Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 2014, 5, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Singh, K.; Almasan, A. Histone H2AX phosphorylation: A marker for DNA damage. Methods Mol. Biol 2012, 920, 613–626. [Google Scholar] [CrossRef]
- Ma, Q.; Qu, Z.; Wang, X.; Qiao, K.; Mangi, N.; Fan, S. EMBRYONIC FLOWER2B, coming from a stable QTL, represses the floral transition in cotton. Int. J. Biol. Macromol. 2020, 163, 1087–1096. [Google Scholar] [CrossRef]
- Cruz, A.B.; Bianchetti, R.E.; Alves, F.R.R.; Purgatto, E.; Peres, L.E.P.; Rossi, M.; Freschi, L. Light, Ethylene and Auxin Signaling Interaction Regulates Carotenoid Biosynthesis During Tomato Fruit Ripening. Front. Plant Sci. 2018, 9, 1370. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Jin, P.; Cui, X.; Gu, L.; Lu, Z.; Xue, Y.; Wei, L.; Qi, J.; Song, X.; Luo, M.; et al. Control of transposon activity by a histone H3K4 demethylase in rice. Proc. Natl. Acad. Sci. USA 2013, 110, 1953–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Genes | Forward Primers | Reverse Primers |
|---|---|---|
| Actin | ATCTGCTGGAAGGTGCTGAG | CCAAGCAGCATGAAGATCAA |
| Cg2g031440 | GGGTCTCCTTAATGAAGCTGAAGAG | TTTCATTATCTGTTCTTCCCGGCAA |
| Cg2g038450 | GAAGCTTTGGAGGGAGGATTAGATG | CCATTGCCCTCCACGATATTTTCTA |
| Cg2g041150 | AAAATAAAAGGCCAAAACCCGTCTG | ACAACTTGCACAGCTTCTATGGTAA |
| Cg3g007850 | TGAAGGAAGATGTGGTCAGTTGATG | GCTGTCTACTATTCGCTTCCTCATC |
| Cg3g014720 | TCAGAACTCTAGCCAGGAAGAAGAA | GAAGAAACCTGGTAGATGTCAAGCA |
| Cg5g003020 | TTACCATGTCTCAGCTTGTTGAACA | ATCAATTTGTTCACCATGTCCCTGA |
| Cg5g005310 | ATCTTGCAGCTAACACAGGAATGAA | TACAAGATTCAATGCCAAGTGTCGA |
| Cg5g006280 | GTCTTGAACATGCTGTGGAAGTAGA | TCTTCTCGTCTTCTTTGGTAGCAAC |
| Cg5g018250 | CAAGCATGGCAAAAGGAGAAGAAAA | GTCTCTGATTTTGATGCTTCTTCGC |
| Cg5g033850 | CAAACGGGAATGCAACATATGTCTC | ACAGCTGCTTTACATGATTCAGACA |
| Cg6g013700 | TTCAATCCCTTACATGTGAAGTGCA | TGTTGGTAGATCAAATTGGGCATCA |
| Cg7g006840 | AAGGTTGGAGGATGTTCTGAAGTTC | GCAGTAGTATGATTTTGCAGCTGTG |
| Cg7g013930 | GGTGAAATTGACATGACCAACAGTG | GCCACTGGAAATGCTTTAAATCTCC |
| Cg8g010020 | TCCTGTGTTCTACCCTACTGAAGAG | CACGCGTAACAAATGTAGAACTGTC |
| Cg2g045510 | AACATGACTGATTGCGAAAAGGTTG | TTGCCACAAAGAGTCTCTGATACAC |
| Cg5g034880 | CCTGTTCTCTCCAGTCTTCTTCAAC | TGCCATAACAACAAAGACTTCCTGA |
| Cg2g002200 | GTTTTGGTGAAAGAGAAGCTAAGCG | AAGTCATTCCAGTTTGTCCTAGCTG |
| Annotation Number | AAs | Mw (kDa) | pI |
|---|---|---|---|
| Cg5g033850 | 789 | 88.59 | 6.70 |
| Cg6g013700 | 1010 | 118.93 | 5.38 |
| Cg2g031440 | 947 | 107.39 | 5.46 |
| Cg5g018250 | 1755 | 193.70 | 8.22 |
| Cg3g014720 | 976 | 111.60 | 5.25 |
| Cg8g010020 | 1259 | 140.86 | 7.50 |
| Cg7g013930 | 1135 | 128.62 | 7.30 |
| Cg3g007850 | 874 | 98.63 | 8.18 |
| Cg7g006840 | 1635 | 181.41 | 5.82 |
| Cg5g005310 | 456 | 53.43 | 5.06 |
| Cg5g006280 | 1666 | 187.79 | 8.67 |
| Cg5g012890 | 926 | 106.20 | 9.03 |
| Cg5g003020 | 1849 | 210.07 | 7.64 |
| Cg2g041150 | 1004 | 114.88 | 8.28 |
| Cg2g038450 | 1048 | 117.76 | 5.83 |
| Cg2g045510 | 1395 | 189,322.74 | 8.55 |
| Cg5g034880 | 518 | 59,185.24 | 5.24 |
| Cg2g002200 | 401 | 45,456.52 | 5.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Fan, R.; Zeng, J. Comprehensive Analysis of Jumonji Domain C Family from Citrus grandis and Expression Profilings in the Exocarps of “Huajuhong” (Citrus grandis “Tomentosa”) during Various Development Stages. Horticulturae 2021, 7, 592. https://doi.org/10.3390/horticulturae7120592
Tian Y, Fan R, Zeng J. Comprehensive Analysis of Jumonji Domain C Family from Citrus grandis and Expression Profilings in the Exocarps of “Huajuhong” (Citrus grandis “Tomentosa”) during Various Development Stages. Horticulturae. 2021; 7(12):592. https://doi.org/10.3390/horticulturae7120592
Chicago/Turabian StyleTian, Yuzhen, Ruiyi Fan, and Jiwu Zeng. 2021. "Comprehensive Analysis of Jumonji Domain C Family from Citrus grandis and Expression Profilings in the Exocarps of “Huajuhong” (Citrus grandis “Tomentosa”) during Various Development Stages" Horticulturae 7, no. 12: 592. https://doi.org/10.3390/horticulturae7120592
APA StyleTian, Y., Fan, R., & Zeng, J. (2021). Comprehensive Analysis of Jumonji Domain C Family from Citrus grandis and Expression Profilings in the Exocarps of “Huajuhong” (Citrus grandis “Tomentosa”) during Various Development Stages. Horticulturae, 7(12), 592. https://doi.org/10.3390/horticulturae7120592






