Glucose Signaling-Mediated Coordination of Cell Growth and Cell Cycle in Saccharomyces Cerevisiae
Abstract
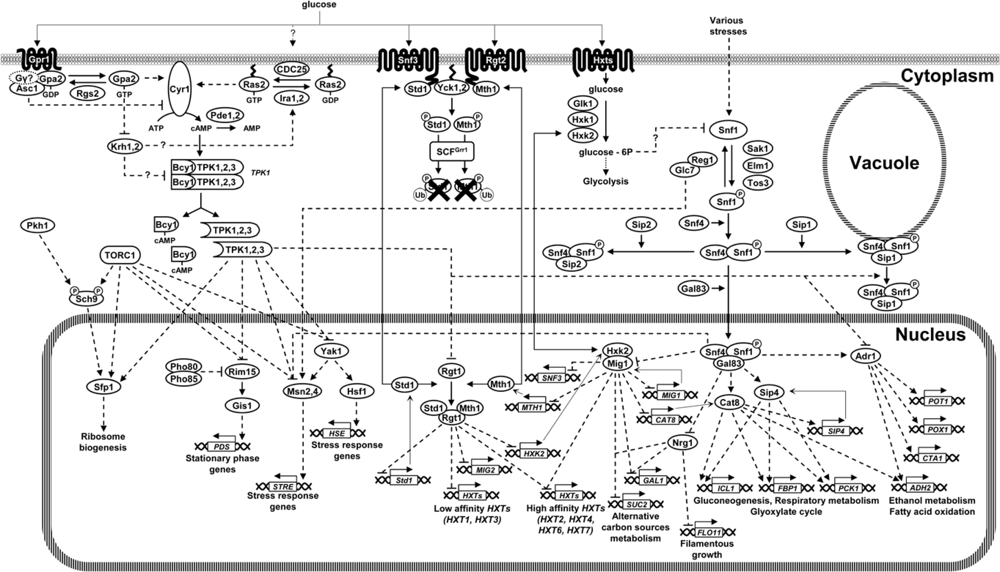
:1. Introduction
2. Glucose Transport in S. Cerevisiae Relies on a Multi-Component Uptake System
3. Sensing Extracellular Glucose
3.1. Glucose Induction Signal: the Snf3-Rgt2 Signaling Transduction Pathway
3.1.1. The Snf3 and Rgt2 Sensors
3.1.2. Downstream Elements of the Snf3-Rgt2 Signaling Transduction Pathway
3.2. The cAMP/PKA Pathway
3.2.1. The GPCR System
3.2.2. Interdependence between the GPCR System, Ras and Glucose Phosphorylation in Glucose-Induced cAMP Signaling
3.2.3. Downstream Elements of the cAMP/PKA Pathway
4. Sensing Intracellular Glucose
4.1. The Snf1 Signaling Transduction Pathway
4.1.1. The Snf1 Protein Kinase Complex and Its Regulation
4.1.2. Downstream Effectors of the Snf1 Protein Kinase Complex: Transcriptional Control in Response to Glucose Limitation
4.1.3. The Role of Snf1 Complex in Cell Cycle regulation
5. Interconnections among Glucose Sensing Mechanisms
5.1. Integrated Regulation of HXT Expression
5.2. Snf1, Rgt2/Snf3 and Glucose Repression
5.3. Glucose Dependent Regulation of HXK2 Expression: Hints for a Possible Cross-Talk among the cAMP/PKA, Rgt2/Snf3 and Snf1 Pathways
6. The Transcriptional Response to Glucose: Contributions of the Diverse Signaling Circuits
6.1. Nutrients, Transcriptional Profile and Growth Rate
7. Connecting Glucose Sensing and Availability to Cell Growth and Division
7.1. PKA Signaling and Cell Growth
7.2. Connections with other Nutrient Sensing Pathways Pathway
7.2.1. The TOR Pathway
7.2.2. TOR-PKA Connections
7.3. Nutritional and Genetic Modulation of Cell Size
8. Conclusions
Acknowledgments
References and Notes
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F; Hoheisel, J.D.; Jacq, C.; Johnston, M.; Louis, E.J.; Mewes, H.W.; Murakami, Y.; Philippsen, P.; Tettelin, H.; Oliver, S.G. Life with 6000 genes. Science 1996, 274, 563–567. [Google Scholar]
- Kitano, H. Looking beyond the details: a rise in system-oriented approaches in genetics and molecular biology. Curr. Genet 2002, 41, 1–10. [Google Scholar]
- Westerhoff, H.V.; Palsson, B.O. The evolution of molecular biology into systems biology. Nat. Biotechno 2004, 22, 1249–1252. [Google Scholar]
- Alberghina, L.; Westerhoff, H.V. Systems Biology, Definitions and Perspectives. In Topics in Current Genetics; Springer Verlag: Berlin, Heidelberg, Germany, 2005; p. 408. [Google Scholar]
- Gray, J.V.; Petsko, G.A.; Johnston, G.C.; Ringe, D.; Singer, R.A.; Werner-Washburne, M. Sleeping beauty: quiescence in Saccharomyces cerevisiae. Microbiol Mol. Biol. Rev 2004, 68, 187–206. [Google Scholar]
- Boles, E.; Hollenberg, C.P. The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev 1997, 1, 85–111. [Google Scholar]
- Ozcan, S.; Johnston, M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev 1999, 63, 554–569. [Google Scholar]
- Reifenberger, E.; Boles, E.; Ciriacy, M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur J. Biochem 1997, 245, 324–333. [Google Scholar]
- Reifenberger, E.; Freidel, K.; Ciriacy, M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol Microbiol 1995, 16, 157–167. [Google Scholar]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Freidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett 1999, 464, 123–128. [Google Scholar]
- Ozcan, S.; Dover, J.; Johnston, M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J 1998, 17, 2566–2573. [Google Scholar]
- Maier, A.; Völker, B.; Boles, E.; Fuhrmann, GF. Characterisation of glucose transport in Saccharomyces cerevisiae with plasma membrane vesicles (countertransport) and intact cells (initial uptake) with single Hxt1, Hxt2, Hxt3, Hxt4, Hxt6, Hxt7 or Gal2 transporters. FEMS Yeast Res 2002, 2, 539–550. [Google Scholar]
- Ozcan, S.; Johnston, M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell Biol 1995, 15, 1564–1572. [Google Scholar]
- Ozcan, S.; Dover, J.; Rosenwald, A.G.; Wölfl, S.; Johnston, M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci USA 1996, 93, 12428–12432. [Google Scholar]
- Johnston, M.; Kim, J.H. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans 2005, 33, 247–252. [Google Scholar]
- Krampe, S.; Stamm, O.; Hollenberg, C.P.; Boles, E. Catabolite inactivation of the high-affinity hexose transporters Hxt6 and Hxt7 of Saccharomyces cerevisiae occurs in the vacuole after internalization by endocytosis. FEBS Lett 1998, 441, 343–347. [Google Scholar]
- Krampe, S.; Boles, E. Starvation-induced degradation of yeast hexose transporter Hxt7p is dependent on endocytosis, autophagy and the terminal sequences of the permease. FEBS Lett 2002, 513, 193–196. [Google Scholar]
- Ye, L.; Kruckeberg, A.L.; Berden, J.A.; van Dam, K. Growth and glucose repression are controlled by glucose transport in Saccharomyces cerevisiae cells containing only one glucose transporter. J. Bacteriol 1999, 181, 4673–4675. [Google Scholar]
- van Suylekom, D.; van Donselaar, E.; Blanchetot, C.; Do Ngoc, L.N.; Humbel, B.M.; Boonstra, J. Degradation of the hexose transporter Hxt5p in Saccharomyces cerevisiae. Biol. Cell 2007, 99, 13–23. [Google Scholar]
- Diderich, J.A.; Schepper, M.; van Hoek, P.; Luttik, MA.; van Dijken, J.P.; Pronk, J.T.; Klaassen, P.; Boelens, H.F.; de Mattos, M.J.; van Dam, K.; Kruckeberg, A.L. Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. J Biol. Chem 1999, 274, 15350–15359. [Google Scholar]
- Zaman, S.; Lippman, S.I.; Schneper, L.; Slonim, N.; Broach, J.R. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol 2009, 5, 245. [Google Scholar]
- Ozcan, S.; Johnston, M. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol. Cell Biol 1996, 16, 5536–5545. [Google Scholar]
- Ye, L.; Berden, J.A.; van Dam, K.; Kruckeberg, A.L. Expression and activity of the Hxt7 high-affinity hexose transporter of Saccharomyces cerevisiae. Yeast 2001, 18, 1257–1267. [Google Scholar]
- Dlugai, S.; Hippler, S.; Wieczorke, R.; Boles, E. Glucose-dependent and -independent signaling functions of the yeast glucose sensor Snf3. FEBS Lett 2001, 505, 389–392. [Google Scholar]
- Schulte, F.; Wieczorke, R.; Hollenberg, C.P.; Boles, E. The HTR1 gene is a dominant negative mutant allele of MTH1 and blocks Snf3- and Rgt2-dependent glucose signaling in yeast. J. Bacteriol 2000, 182, 540–542. [Google Scholar]
- Liang, H; Gaber, RF. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol. Biol. Cell 1996, 7, 1953–66. [Google Scholar]
- Diderich, J.A.; Schuurmans, J.M.; Van Gaalen, M.C.; Kruckeberg, A.L.; Van Dam, K. Functional analysis of the hexose transporter homologue HXT5 in Saccharomyces cerevisiae. Yeast 2001, 16, 1515–1524. [Google Scholar]
- Verwaal, R.; Paalman, J.W.; Hogenkamp, A.; Verkleij, A.J.; Verrips, C.T.; Boonstra, J. HXT5 expression is determined by growth rates in Saccharomyces cerevisiae. Yeast 2002, 19, 1029–38. [Google Scholar]
- Verwaal, R.; Arako, M.; Kapur, R.; Verkleij, A.J.; Verrips, C.T.; Boonstra, J. HXT5 expression is under control of STRE and HAP elements in the HXT5 promoter. Yeast 2004, 21, 747–757. [Google Scholar]
- Greatrix, B.W.; van Vuuren, H.J. Expression of the HXT13, HXT15 and HXT17 genes in Saccharomyces cerevisiae and stabilization of the HXT1 gene transcript by sugar-induced osmotic stress. Curr Genet 2006, 49, 205–217. [Google Scholar]
- Ozcan, S.; Leong, T.; Johnston, M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell Biol 1996, 16, 6419–6426. [Google Scholar]
- Kim, J.H.; Brachet, V.; Moriya, H.; Johnston, M. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot Cell 2006, 5, 167–173. [Google Scholar]
- Kim, J.H.; Johnston, M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J. Biol. Chem 2006, 281, 26144–26149. [Google Scholar]
- Kaniak, A.; Xue, Z.; Macool, D.; Kim, J.H.; Johnston, M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell 2004, 3, 221–231. [Google Scholar]
- Hirayama, T.; Maeda, T.; Saito, H.; Shinozaki, K. Cloning and characterization of seven cDNAs for hyperosmolarity-responsive (HOR) genes of Saccharomyces cerevisiae. Mol. Gen Genet 1995, 249, 127–138. [Google Scholar]
- Tomás-Cobos, L.; Casadomé, L.; Mas, G.; Sanz, P.; Posas, F. Expression of the HXT1 low affinity glucose transporter requires the coordinated activities of the HOG and glucose signalling pathways. J. Biol. Chem 2004, 279, 22010–22019. [Google Scholar]
- Tomás-Cobos, L.; Viana, R.; Sanz, P. TOR kinase pathway and 14-3-3 proteins regulate glucose-induced expression of HXT1, a yeast low-affinity glucose transporter. Yeast 2005, 22, 471–479. [Google Scholar]
- Tomás-Cobos, L.; Sanz, P. Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Bio. Chem. J 2002, 368, 657–663. [Google Scholar]
- Petit, T.; Diderich, J.A.; Kruckeberg, A.L.; Gancedo, C.; Van Dam, K. Hexokinase regulates kinetics of glucose transport and expression of genes encoding hexose transporters in Saccharomyces cerevisiae. J. Bacteriol 2000, 182, 6815–6818. [Google Scholar]
- Belinchón, M.M.; Gancedo, J.M. Different signalling pathways mediate glucose induction of SUC2, HXT1 and pyruvate decarboxylase in yeast. FEMS Yeast Res 2007, 7, 40–47. [Google Scholar]
- Belinchon, M.M.; Gancedo, J.M. Glucose controls multiple processes in Saccharomyces cerevisiae through diverse combinations of signaling pathways. FEMS Yeast Res 2007, 7, 808–818. [Google Scholar]
- Coons, D.M.; Vagnoli, P.; Bisson, L.F. The C-terminal domain of Snf3p is sufficient to complement the growth defect of snf3 null mutations in Saccharomyces cerevisiae: SNF3 functions in glucose recognition. Yeast 1997, 13, 9–20. [Google Scholar]
- Vagnoli, P.; Coons, D.M.; Bisson, L.F. The C-terminal domain of Snf3p mediates glucose-responsive signal transduction in Saccharomyces cerevisiae. FEMS Microbiol Lett 1998, 160, 31–36. [Google Scholar]
- Moriya, H.; Johnston, M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 2004, 101, 1572–1577. [Google Scholar]
- Ozcan, S. Two different signals regulate repression and induction of gene expression by glucose. J. Biol. Chem 2002, 277, 46993–46997. [Google Scholar]
- Dietvorst, J.; Karhumaa, K.; Kielland-Brandt, M.C.; Brandt, A. Amino acid residues involved in ligand preference of the Snf3 transporter-like sensor in Saccharomyces cerevisiae. Yeast 2010, 27, 131–138. [Google Scholar]
- Kim, J.H.; Polish, J.; Johnston, M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol. Cell Biol 2003, 23, 5208–5216. [Google Scholar]
- Kim, J.H. DNA-binding properties of the yeast Rgt1 repressor. Biochimie 2009, 91, 300–303. [Google Scholar]
- Mosley, A.L.; Lakshmanan, J.; Aryal, B.K.; Ozcan, S. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem 2003, 278, 10322–10327. [Google Scholar]
- Flick, K.M.; Spielewoy, N.; Kalashnikova, T.I.; Guaderrama, M.; Zhu, Q.; Chang, H.C.; Wittenberg, C. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 2003, 8, 3230–3241. [Google Scholar]
- Polish, J.A.; Kim, J.H.; Johnston, M. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics 2005, 169, 583–594. [Google Scholar]
- Schmidt, M.C.; McCartney, R.R.; Zhang, X.; Tillman, T.S.; Solimeo, H.; Wölfl, S.; Almonte, C.; Watkins, S.C. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell Biol 1999, 19, 4561–4571. [Google Scholar]
- Lafuente, M.J.; Gancedo, C.; Jauniaux, J.C.; Gancedo, J.M. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol 2000, 35, 161–72. [Google Scholar]
- Lakshmanan, J.; Mosley, A.L.; Ozcan, S. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr. Genet 2003, 44, 19–25. [Google Scholar]
- Sabina, J.; Johnston, M. Asymmetric signal transduction through paralogs that comprise a genetic switch for sugar sensing in S. cerevisiae. J. Biol. Chem 2009, 284, 29635–29643. [Google Scholar]
- Pasula, S.; Jouandot., D.; Kim, J.H. Biochemical evidence for glucose-independent induction of HXT expression in Saccharomyces cerevisiae. FEBS Lett 2007, 581, 3230–3234. [Google Scholar]
- Spielewoy, N.; Flick, K.; Kalashnikova, T.I.; Walker, J.R.; Wittenberg, C. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell Biol 2004, 24, 8994–9005. [Google Scholar]
- Pasula, S.; Chakraborty, S.; Choi, J.H.; Kim, J.H. Role of casein kinase 1 in the glucose sensor-mediated signaling pathway in yeast. BMC Cell Biol 2010, 11, 17. [Google Scholar]
- Zaman, S.; Lippman, S.I.; Zhao, X.; Broach, J.R. How Saccharomyces responds to nutrients. Annu. Rev. Genet 2008, 4, 27–81. [Google Scholar]
- Santangelo, G.M. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol. Biol. Rev 2006, 70, 253–282. [Google Scholar]
- Thevelein, J.M.; de Winde, J.H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol 1999, 33, 904–918. [Google Scholar]
- Rolland, F.; Winderickx, J.; Thevelein, J.M. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res 2002, 2, 183–201. [Google Scholar]
- Martegani, E.; Baroni, M.D.; Frascotti, G.; Alberghina, L. Molecular cloning and transcriptional analysis of the start gene CDC25 of Saccharomyces cerevisiae. EMBO J 1986, 5, 2363–2369. [Google Scholar]
- Damak, F.; Boy-Marcotte, E.; Le-Roscouet, D.; Guilbaud, R.; Jacquet, M. SDC25, a CDC25-like gene which contains a RAS-activating domain and is a dispensable gene of Saccharomyces cerevisiae. Mol. Cell Biol 1991, 11, 202–212. [Google Scholar]
- Tanaka, K.; Nakafuku, M.; Satoh, T.; Marshall, M.S.; Gibbs, J.B.; Matsumoto, K.; Kaziro, Y.; Toh-e, A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell 1990, 60, 803–807. [Google Scholar]
- Tanaka, K.; Nakafuku, M.; Tamanoi, F.; Kaziro, Y.; Matsumoto, K.; Toh-e, A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol. Cell Biol 1990, 10, 4303–4313. [Google Scholar]
- Toda, T.; Uno, I.; Ishikawa, T.; Powers, S.; Kataoka, T.; Broek, D.; Cameron, S.; Broach, J.; Matsumoto, K.; Wigler, M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 1985, 40, 27–36. [Google Scholar]
- Casperson, G.F.; Walker, N.; Bourne, H.R. Isolation of the gene encoding adenylate cyclase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1985, 82, 5060–5063. [Google Scholar]
- Sass, P.; Field, J.; Nikawa, J.; Toda, T.; Wigler, M. Cloning and characterization f the high-affinity cAMP phosphodiesterase of S. cerevisiae. Proc. Natl. Acad. Sci. USA 1986, 83, 9303–9307. [Google Scholar]
- Nikawa, J.; Sass, P.; Wigler, M. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol. Cell Biol 1987, 7, 3629–3636. [Google Scholar]
- Colombo, S.; Ma, P.; Cauwenberg, L.; Winderickx, J.; Crauwels, M.; Teunissen, A.; Nauwelaers, D.; de Winde, J.H.; Gorwa, M.F.; Colavizza, D.; Thevelein, J.M. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J 1998, 17, 3326–3341. [Google Scholar]
- Rudoni, S.; Colombo, S.; Coccetti, P.; Martegani, E. Role of guanine nucleotides in the regulation of the Ras/cAMP pathway in Saccharomyces cerevisiae. Biochim. Biophys. Acta 2001, 1538, 181–189. [Google Scholar]
- Colombo, S.; Ronchetti, D.; Thevelein, J.M.; Winderickx, J.; Martegani, E. Activation state of the Ras2 protein and glucose-induced signaling in Saccharomyces cerevisiae. J. Biol. Chem 2004, 279, 46715–46722. [Google Scholar]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Wigler, M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 1987, 50, 277–287. [Google Scholar]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Scott, J.D.; McMullen, B.; Hurwitz, M.; Krebs, E.G.; Wigler, M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell Biol 1987, 7, 1371–1377. [Google Scholar]
- Robertson, L.S.; Fink, G.R. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl Acad. Sci. USA 1998, 95, 13783–13787. [Google Scholar]
- Robertson, L.S.; Causton, H.C.; Young, R.A.; Fink, G.R. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl Acad. Sci. USA 2000, 97, 5984–5988. [Google Scholar]
- Pan, X.; Heitman, J. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol 2002, 22, 3981–3993. [Google Scholar]
- Chevtzoff, C.; Vallortigara, J.; Averet, N.; Rigoulet, M.; Devin, A. The yeastcAMPprotein kinase Tpk3p is involved in the regulation of mitochondrial enzymatic content during growth. Biochim. Biophys. Acta 2005, 1706, 117–125. [Google Scholar]
- Palomino, A.; Herrero, P.; Moreno, F. Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res 2006, 34, 1427–38. [Google Scholar]
- Griffioen, G.; Anghileri, P.; Imre, E.; Baroni, M.D.; Ruis, H. Nutritional control of nucleocytoplasmic localization of cAMP-dependent protein kinase catalytic and regulatory subunits in Saccharomyces cerevisiae. J. Biol. Chem 2000, 275, 449–456. [Google Scholar]
- Griffioen, G.; Branduardi, P.; Ballarini, A.; Anghileri, P.; Norbeck, J.; Baroni, M.D.; Ruis, H. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol Cell Biol 2001, 21, 511–523. [Google Scholar]
- Griffioen, G.; Thevelein, J.M. Molecular mechanisms controlling the localisation of protein kinase A. Curr Genet 2002, 41, 199–207. [Google Scholar]
- Schmelzle, T.; Beck, T.; Martin, D.E.; Hall, M.N. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell Biol 2004, 24, 338–351. [Google Scholar]
- Gancedo, J.M. The early steps of glucose signalling in yeast. FEMS Microbiol Rev 2008, 32, 673–704. [Google Scholar]
- Xue, Y.; Batlle, M.; Hirsch, J.P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J 1998, 17, 1996–2007. [Google Scholar]
- Kraakman, L.; Lemaire, K.; Ma, P.; Teunissen, A.W.; Donaton, M.C.; Van Dijck, P.; Winderickx, J.; de Winde, J.H.; Thevelein, J.M. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol 1999, 32, 1002–1012. [Google Scholar]
- Nakafuku, M.; Obara, T.; Kaibuchi, K.; Miyajima, I.; Miyajima, A.; Itoh, H.; Nakamura, S.; Arai, K.; Matsumoto, K.; Kaziro, Y. Isolation of a second yeast Saccharomyces cerevisiae gene (GPA2) coding for guanine nucleotide-binding regulatory protein: studies on its structure and possible functions. Proc. Natl. Acad. Sci. USA 1988, 85, 1374–1378. [Google Scholar]
- Rolland, F.; De Winde, J.H.; Lemaire, K.; Boles, E.; Thevelein, J.M.; Winderickx, J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol 2000, 38, 348–358. [Google Scholar]
- Lemaire, K.; Van de Velde, S.; Van Dijck, P.; Thevelein, J.M. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 2004, 16, 293–299. [Google Scholar]
- Rubio-Texeira, M.; Van Zeebroeck, G.; Voordeckers, K.; Thevelein, J.M. Saccharomyces cerevisiae plasma membrane nutrient sensors and their role in PKA signalling. FEMS Yeast Res 2010, 10, 134–149. [Google Scholar]
- Johnston, M.; Carlson, M. The Molecular and Cellular Biology of the Yeast Saccharomyces; Gene Expression Jones, P.E., Broach, J.R., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1993; pp. 193–282. [Google Scholar]
- Alberghina, L.; Rossi, R.L.; Querin, L.; Wanke, V.; Vanoni, M. A cell sizer network involving Cln3 and Far1 controls entrance into S phase in the mitotic cycle of budding yeast. J. Cell Biol 2004, 167, 433–443. [Google Scholar]
- Tamaki, H.; Yun, C.W.; Mizutani, T.; Tsuzuki, T.; Takagi, Y.; Shinozaki, M.; Kodama, Y.; Shirahige, K.; Kumagai, H. Glucose-dependent cell size is regulated by a G protein-coupled receptor system in yeast Saccharomyces cerevisiae. Genes Cell 2005, 10, 193–206. [Google Scholar]
- Tamaki, H. Glucose-stimulated cAMP-protein kinase A pathway in yeast Saccharomyces cerevisiae. J. Biosci. Bioeng 2007, 104, 245–250. [Google Scholar]
- Zeller, C.E.; Parnell, S.C.; Dohlman, H.G. The RACK1 ortholog Asc1 functions as a G-protein beta subunit coupled to glucose responsiveness in yeast. J. Biol. Chem 2007, 282, 25168–25176. [Google Scholar]
- Harashima, T.; Heitman, J. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol. Cell 2002, 10, 163–173. [Google Scholar]
- Harashima, T.; Heitman, J. Galpha subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol. Biol. Cell 2005, 16, 4557–4571. [Google Scholar]
- Lu, A.; Hirsch, J.P. Cyclic AMP-independent regulation of protein kinase A substrate phosphorylation by Kelch repeat proteins. Eukaryot Cell 2005, 4, 1794–1800. [Google Scholar]
- Harashima, T.; Anderson, S.; Yates, J.R., 3rd; Heitman, J. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol. Cell 2006, 22, 819–830. [Google Scholar]
- Peeters, T.; Louwet, W.; Geladé, R.; Nauwelaers, D.; Thevelein, J.M.; Versele, M. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. USA 2006, 103, 13034–13039. [Google Scholar]
- Peeters, T; Versele, M; Thevelein, JM. Directly from Galpha to protein kinase A: the kelch repeat protein bypass of adenylate cyclase. Trends Biochem. Sci 2007, 32, 547–554. [Google Scholar]
- Versele, M.; de Winde, J.H.; Thevelein, J.M. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J 1999, 18, 5577–5591. [Google Scholar]
- Rolland, F.; Wanke, V.; Cauwenberg, L.; Ma, P.; Boles, E.; Vanoni, M.; de Winde, J.H.; Thevelein, J.M.; Winderickx, J. The role of hexose transport and phosphorylation in cAMP signalling in the yeast Saccharomyces cerevisiae. FEMS Yeast Res 2001, 1, 33–45. [Google Scholar]
- Goldberg, D.; Segal, M.; Levitzki, A. Cdc25 is not the signal receiver for glucose induced cAMP response in S. cerevisiae. FEBS Lett 1994, 356, 249–254. [Google Scholar]
- Mbonyi, K.; Beullens, M.; Detremerie, K.; Geerts, L.; Thevelein, J.M. Requirement of one functional RAS gene and inability of an oncogenic ras-variant to mediate the glucose-induced cAMP signal in the yeast Saccharomyces cerevisiae. Mol. Cell Biol 1988, 8, 3051–3057. [Google Scholar]
- Munder, T.; Kuntzel, H. Glucose-induced cAMP signaling in Saccharomyces cerevisiae is mediated by the CDC25 protein. FEBS Lett 1989, 242, 341–345. [Google Scholar]
- Van Aelst, L.; Boy-Marcotte, E.; Camonis, J.H.; Thevelein, J.M.; Jacquet, M. The C-terminal part of the CDC25 gene product plays a key role in signal transduction in the glucose-induced modulation of cAMP level in Saccharomyces cerevisiae. Eur. J. Biochem 1990, 193, 675–680. [Google Scholar]
- Van Aelst, L.; Jans, A.W.H.; Thevelein, J.M. Involvement of the CDC25 gene product in the signal transmission pathway of the glucose-induced RAS-mediated cAMP signal in the yeast Saccharomyces cerevisiae. J. Gen Microbiol 1991, 137, 341–349. [Google Scholar]
- Bhattacharya, S.; Chen, L.; Broach, J.R.; Powers, S. Ras membrane targeting is essential for glucose signaling but not for viability in yeast. Proc. Natl. Acad. Sci. USA 1995, 92, 2984–2988. [Google Scholar]
- Wang, Y.; Pierce, M.; Schneper, L.; Güldal, C.G.; Zhang, X.; Tavazoie, S.; Broach, J.R. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol 2004, 2, E128. [Google Scholar]
- Slattery, M.G.; Liko, D.; Heideman, W. Protein kinase A, TOR, and glucose transport control the response to nutrient repletion in Saccharomyces cerevisiae. Eukaryot Cell 2008, 7, 358–367. [Google Scholar]
- Jorgensen, P.; Rupes, I.; Sharom, J.R.; Schneper, L.; Broach, J.R.; Tyers, M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 2004, 18, 2491–2505. [Google Scholar]
- Klein, C.; Struhl, K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell Biol 1994, 14, 1920–1928. [Google Scholar]
- Neuman-Silberberg, F.S.; Bhattacharya, S.; Broach, J.R. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol. Cell Biol 1995, 15, 3187–3196. [Google Scholar]
- Zurita-Martinez, S.A.; Cardenas, M.E. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot Cell 2005, 4, 63–71. [Google Scholar]
- Marion, R.M.; Regev., A.; Segal, E.; Barash, Y.; Koller, D.; Friedman, N.; O’Shea, E.K. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA 2004, 101, 14315–14322. [Google Scholar]
- Fingerman, I.; Nagaraj, V.; Norris, D.; Vershon, A.K. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot Cell 2003, 2, 1061–1068. [Google Scholar]
- Martínez-Pastor, M.T.; Marchler, G.; Schüller, C.; Marchler-Bauer, A.; Ruis, H.; Estruch, F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 1996, 15, 2227–2235. [Google Scholar]
- Görner, W.; Durchschlag, E.; Martinez-Pastor, M.T.; Estruch, F.; Ammerer, G.; Hamilton, B.; Ruis, H.; Schüller, C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev 1998, 12, 586–597. [Google Scholar]
- Smith, A.; Ward, M.P.; Garrett, S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J 1998, 17, 3556–3564. [Google Scholar]
- Boy-Marcotte, E.; Perrot, M.; Bussereau, F.; Boucherie, H.; Jacquet, M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol 1998, 180, 1044–1052. [Google Scholar]
- Moskvina, E.; Schuller, C.T.; Maurer, C.; Mager, W.H.; Ruis, H. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 1998, 14, 1041–1050. [Google Scholar]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar]
- Causton, H.C.; Ren, B.; Koh, S.S.; Harbison, C.T.; Kanin, E.; Jennings, E.G.; Lee, T.I.; True, H.L.; Lander, E.S.; Young, R.A. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001, 12(2), 323–337. [Google Scholar]
- Görner, W.; Durchschlag, E.; Wolf, J.; Brown, E.L.; Ammerer, G.; Ruis, H.; Schüller, C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J 2002, 21, 135–144. [Google Scholar]
- Santhanam, A.; Hartley, A.; Duvel, K.; Broach, J.R.; Garrett, S. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot. Cell 2004, 3, 1261–1271. [Google Scholar]
- Beck, T.; Hall, M.N. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 1999, 402, 689–692. [Google Scholar]
- Hirata, Y.; Andoh, T.; Asahara, T.; Kikuchi, A. Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol. Biol. Cell 2003, 14, 302–312. [Google Scholar]
- Boy-Marcotte, E.; Garmendia, C.; Garreau, H.; Lallet, S.; Mallet, L.; Jacquet, M. The transcriptional activation region of Msn2p, in Saccharomyces cerevisiae, is regulated by stress but is insensitive to the cAMP signalling pathway. Mol. Genet. Genomics 2006, 275, 277–287. [Google Scholar]
- Durchschlag, E.; Reiter, W.; Ammerer, G.; Schuller, C. Nuclear localization destabilizes the stressregulated transcription factor Msn2. J. Biol. Chem 2004, 279, 55425–55432. [Google Scholar]
- Lallet, S.; Garreau, H.; Poisier, C.; Boy-Marcotte, E.; Jacquet, M. Heat shock-induced degradation of Msn2p, a Saccharomyces cerevisiae transcription factor, occurs in the nucleus. Mol. Genet. Genomics 2004, 272, 353–362. [Google Scholar]
- Lallet., S; Garreau, H.; Garmendia-Torres, C.; Szestakowska, D.; Boy-Marcotte, E.; Quevillon-Chéruel, S.; Jacquet, M. Role of Gal11, a component of the RNA polymerase II mediator in stress-induced hyperphosphorylation of Msn2 in Saccharomyces cerevisiae. Mol. Microbiol 2006, 62, 438–452. [Google Scholar]
- Bose, S.; Dutko, J.A.; Zitomer, R.S. Genetic factors that regulate the attenuation of the general stress response of yeast. Genetics 2005, 169, 1215–1226. [Google Scholar]
- Lee, P.; Cho, B.R.; Joo, H.S.; Hahn, J.S. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol 2008, 70, 882–895. [Google Scholar]
- Garrett, S.; Menold, M.M.; Broach, J.R. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol. Cell Biol 1991, 11, 4045–4052. [Google Scholar]
- Garrett, S.; Broach, J. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP dependent protein kinase. Genes Dev 1989, 9, 1336–1348. [Google Scholar]
- Ward, M.P.; Garrett, S. Suppression of a yeast cyclic AMP-dependent protein kinase defect by overexpression of SOK1, a yeast gene exhibiting sequence similarity to a developmentally regulated mouse gene. Mol. Cell Biol 1994, 14, 5619–5627. [Google Scholar]
- Moriya, H.; Shimizu-Yoshida, Y.; Omori, A.; Iwashita, S.; Katoh, M.; Sakai, A. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev 2001, 15, 1217–1228. [Google Scholar]
- Martin, D.E.; Soulard, A.; Hall, M.N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 2004, 119, 969–979. [Google Scholar]
- Ferguson, S.B.; Anderson, E.S.; Harshaw, R.B.; Thate, T.; Craig, N.L.; Nelson, H.C. Protein kinase A regulates constitutive expression of small heat-shock genes in an Msn2/4p-independent and Hsf1p-dependent manner in Saccharomyces cerevisiae. Genetics 2005, 169, 1203–1214. [Google Scholar]
- Hahn, J.S.; Hu, Z.; Thiele, D.J.; Iyer, V.R. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell Biol 2004, 24, 5249–5256. [Google Scholar]
- Eastmond, D.L.; Nelson, H.C. Genome-wide analysis reveals new roles for the activation domains of the Saccharomyces cerevisiae heat shock transcription factor (Hsf1) during the transient heat shock response. J Biol Chem 2006, 281, 32909–32921. [Google Scholar]
- Wiederrecht, G.; Seto, D.; Parker, C.S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 1988, 54, 841–853. [Google Scholar]
- Smith, B.J.; Yaffe, M.P. A mutation in the yeast heat-shock factor gene causes temperature-sensitive defects in both mitochondrial protein import and the cell cycle. Mol. Cell Biol 1991, 11, 2647–2655. [Google Scholar]
- Zarzov, P.; Boucherie, H.; Mann, C. A yeast heat shock transcription factor (Hsf1) mutant is defective in both Hsc82/Hsp82 synthesis and spindle pole body duplication. J. Cell Sci 1997, 110, 1879–1891. [Google Scholar]
- Imazu, H.; Sakurai, H. Saccharomyces cerevisiae heat shock transcription factor regulates cell wall remodeling in response to heat shock. Eukaryot Cell 2005, 4, 1050–1056. [Google Scholar]
- Hahn, J.S.; Thiele, D.J. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J. Biol. Chem 2004, 279, 5169–5176. [Google Scholar]
- Boy-Marcotte, E.; Lagniel, G.; Perrot, M.; Bussereau, F.; Boudsocq, A.; Jacquet, M.; Labarre, J. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol 1999, 33, 274–283. [Google Scholar]
- Amorós, M.; Estruch, F. Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol. Microbiol 2001, 39, 1523–1532. [Google Scholar]
- Treger, J.M.; Schmitt, A.P.; Simon, J.R.; McEntee, K. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J. Biol. Chem 1998, 273, 26875–26879. [Google Scholar]
- Yamamoto, N.; Maeda, Y.; Ikeda, A.; Sakurai, H. Regulation of thermotolerance by stress-induced transcription factors in Saccharomyces cerevisiae. Eukaryot Cell 2008, 7, 783–790. [Google Scholar]
- Reinders, A.; Burckert, N.; Boller, T.; Wiemken, A.; De Virgilio, C. Saccharomyces cerevisiae cAMPdependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev 1998, 12, 2943–2955. [Google Scholar]
- Swinnen, E.; Wanke, V.; Roosen, J.; Smets, B.; Dubouloz, F.; Pedruzzi, I.; Cameroni, E.; De Virgilio, C.; Winderickx, J. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div 2006, 3, 1–3. [Google Scholar]
- Pedruzzi, I.; Dubouloz, F.; Cameroni, E.; Wanke, V.; Roosen, J.; Winderickx, J.; De Virgilio, C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 2003, 12, 1607–1613. [Google Scholar]
- Wanke, V.; Pedruzzi, I.; Cameroni, E.; Dubouloz, F.; De Virgilio, C. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J 2005, 24, 4271–4278. [Google Scholar]
- Roosen, J.; Engelen, K.; Marchal, K.; Mathys, J.; Griffioen, G.; Cameroni, E.; Thevelein, J.M.; De Virgilio, C.; De Moor, B.; Winderickx, J. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol. Microbiol 2005, 55, 862–880. [Google Scholar]
- Pedruzzi., I.; Bürckert, N.; Egger, P.; De Virgilio, C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J 2000, 19, 2569–2579. [Google Scholar]
- Cameroni, E.; Hulo, N.; Roosen, J.; Winderickx, J.; De Virgilio, C. The novel yeast PAS kinase Rim 15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle 2004, 3, 462–468. [Google Scholar]
- Vanoni, M.; Sollitti, P.; Goldenthal, M.; Marmur, J. Structure and regulation of the multigene family controlling maltose fermentation in budding yeast. Prog. Nucleic Acid Res. Mol. Biol 1989, 37, 281–322. [Google Scholar]
- Carlson, M. Glucose repression in yeast. Curr. Opin. Microbiol 1999, 2, 202–207. [Google Scholar]
- Hedbacker, K.; Carlson, M. SNF1/AMPK pathways in yeast. Front Biosci 2008, 13, 2408–2420. [Google Scholar]
- De Winde, J.H.; Crauwels, M.; Hohmann, S.; Thevelein, J.M.; Winderickx, J. Differential requirement of the yeast sugar kinases for sugar sensing in establishing the catabolite-repressed state. Eur. J. Biochem 1996, 241, 633–643. [Google Scholar]
- Ahuatzi, D.; Herrero, P.; de la Cera, T.; Moreno, F. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem 2004, 279, 14440–14446. [Google Scholar]
- Ahuatzi, D.; Riera, A.; Peláez, R.; Herrero, P.; Moreno, F. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem 2007, 282, 4485–4493. [Google Scholar]
- Moreno, F.; Ahuatzi, D.; Riera, A.; Palomino, C.A.; Herrero, P. Glucose sensing through the Hxk2-dependent signalling pathway. Biochem. Soc. Trans 2005, 33, 265–268. [Google Scholar]
- Elbing, K.; Ståhlberg, A.; Hohmann, S.; Gustafsson, L. Transcriptional responses to glucose at different glycolytic rates in Saccharomyces cerevisiae. Eur. J. Biochem 2004, 271, 4855–4864. [Google Scholar]
- Otterstedt, K.; Larsson, C.; Bill, R.M.; Ståhlberg, A.; Boles, E.; Hohmann, S.; Gustafsson, L. Switching the mode of metabolism in the yeast Saccharomyces cerevisiae. EMBO Rep 2004, 5, 532–537. [Google Scholar]
- Mitchelhill, K.I.; Stapleton, D.; Gao, G.; House, C.; Michell, B.; Katsis, F.; Witters, L.A.; Kemp, B.E. MammalianAMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J. Biol. Chem 1994, 269, 2361–2364. [Google Scholar]
- Lo, W.; Duggan, S.L.; Emre, N.C.; Belotserkovskya, R.; Lane, W.S. Shiekhattar R, Berger SL. Snf1-a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 2001, 293, 1142–1146. [Google Scholar]
- Vincent, O.; Townley, R.; Kuchin, S.; Carlson, M. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev 2001, 15, 1104–1114. [Google Scholar]
- Momcilovic, M.; Iram, S.H.; Liu, Y.; Carlson, M. Roles of the glycogen-binding domain and Snf4 in glucose inhibition of SNF1 protein kinase. J. Biol. Chem 2008, 283, 19521–19529. [Google Scholar]
- Wilson, W.A.; Hawley, S.A.; Hardie, D.G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP: ATP ratio. Curr. Biol 1996, 6, 1426–1434. [Google Scholar]
- Johnston, M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet 1999, 15, 29–33. [Google Scholar]
- Nath, N.; McCartney, R.R.; Schmidt, M.C. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol 2003, 23, 3909–3917. [Google Scholar]
- Sutherland, C.M.; Hawley, S.A.; McCartney, R.R.; Leech, A.; Stark, M.J.; Schmidt, M.C.; Hardie, D.G. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol 2003, 13, 1299–1305. [Google Scholar]
- Hong, S.P; Leiper, F.C.; Woods, A.; Carling, D.; Carlson, M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 2003, 100, 8839–8843. [Google Scholar]
- Jiang, R.; Carlson, M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev 1996, 10, 3105–3115. [Google Scholar]
- Ludin, K.; Jiang, R.; Carlson, M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1998, 95, 6245–6250. [Google Scholar]
- Hedbacker, K.; Hong, S.P.; Carlson, M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell Biol 2004, 24, 8255–8263. [Google Scholar]
- Kim, M.D.; Hong, S.P.; Carlson, M. Role of Tos3, a Snf1 protein kinase kinase, during growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Eukaryot Cell 2005, 4, 861–866. [Google Scholar]
- Tu, J.; Carlson, M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J 1995, 14, 5939–5946. [Google Scholar]
- Sanz, P.; Alms, G.R.; Haystead, T.A.; Carlson, M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell Biol 2000, 20, 1321–1328. [Google Scholar]
- McCartney, R.R.; Schmidt, M.C. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem 2001, 276, 36460–36466. [Google Scholar]
- Rubenstein, E.M.; McCartney, R.R.; Zhang, C.; Shokat, K.M.; Shirra, M.K.; Arndt, K.M.; Schmidt, M.C. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem 2008, 283, 222–230. [Google Scholar]
- Hedbacker, K.; Carlson, M. Regulation of the nucleocytoplasmic distribution of Snf1-Gal83 protein kinase. Eukaryot Cell 2006, 5, 1950–1956. [Google Scholar]
- Young, E.T.; Dombek, K.M.; Tachibana, C.; Ideker, T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem 2003, 278, 26146–26158. [Google Scholar]
- Tachibana, C.; Yoo, J.Y.; Tagne, J.B.; Kacherovsky, N.; Lee, T.I.; Young, E.T. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol 2005, 25, 2138–2146. [Google Scholar]
- Usaite, R.; Jewett, M.C.; Oliveira, A.P.; Yates, J.R., 3rd; Olsson, L.; Nielsen, J. Reconstruction of the yeast Snf1 kinase regulatory network reveals its role as a global energy regulator. Mol. Syst. Biol 2009, 5, 319. [Google Scholar]
- Treitel, M.A.; Carlson, M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. USA 1995, 92, 3132–3136. [Google Scholar]
- Lutfiyya, L.L.; Iyer, V.R.; DeRisi, J.; DeVit, M.J.; Brown, P.O.; Johnston, M. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 1998, 150, 1377–1391. [Google Scholar]
- Lutfiyya, L.L.; Johnston, M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell Biol 1996, 16, 4790–4797. [Google Scholar]
- Westholm, J.O.; Nordberg, N.; Murén, E.; Ameur, A.; Komorowski, J.; Ronne, H. Combinatorial control of gene expression by the three yeast repressors Mig1, Mig2 and Mig3. BMC Genomics 2008, 9, 601. [Google Scholar]
- Tzamarias, S.; Struhl, K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev 1995, 9, 821–831. [Google Scholar]
- De Vit, M.J.; Waddle, J.A.; Johnston, M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 1997, 8, 1603–1618. [Google Scholar]
- De Vit, M.J.; Johnston, M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol 1999, 9, 1231–1241. [Google Scholar]
- Rodríguez, A.; De La Cera, T.; Herrero, P.; Moreno, F. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem J 2001, 355, 625–31. [Google Scholar]
- Young, E.T.; Kacherovsky, N.; Van Riper, K. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J Biol Chem 2002, 277, 38095–38103. [Google Scholar]
- Dombek, K.M.; Kacherovsky, N.; Young, E.T. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J. Biol. Chem 2004, 279, 39165–39174. [Google Scholar]
- Hedges, D.; Proft, M.; Entian, K.D. CAT8, a new zinc clusterencoding gene necessary for depression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell Biol 1995, 15, 1915–1922. [Google Scholar]
- Lesage, P.; Yang, X.; Carlson, M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol. Cell Biol 1996, 16, 1921–1928. [Google Scholar]
- Randez-Gil, F.; Bojunga, N.; Proft, M.; Entian, K.D. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol. Cell Biol 1997, 17, 2502–2510. [Google Scholar]
- Vincent, O.; Carlson, M. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J 1998, 17, 7002–7008. [Google Scholar]
- Charbon, G.; Breunig, K.D.; Wattiez, R.; Vandenhaute, J.; Noel-Georis, I. Key role of Ser562/661 in Snf1-dependent regulation of Cat8p in Saccharomyces cerevisiae and Kluyveromyces lactis. Mol. Cell Biol 2004, 24, 4083–4091. [Google Scholar]
- Vincent, O.; Carlson, M. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J 1999, 18, 6672–6681. [Google Scholar]
- Schmidt, M.C.; McCartney, R.R. beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J 2000, 19, 4936–4943. [Google Scholar]
- Mayordomo, I.; Estruch, F.; Sanz, P. Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (Stress Response Element)-regulated genes. J. Biol. Chem 2002, 277, 35650–35656. [Google Scholar]
- De Wever, V.; Reiter, W.; Ballarini, A.; Ammerer, G.; Brocard, C. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J 2005, 24, 4115–4123. [Google Scholar]
- Bertram, P.G.; Choi, J.H.; Carvalho, J.; Chan, T.F.; Ai, W.; Zheng, X.F. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol. Cell Biol 2002, 22, 1246–1252. [Google Scholar]
- Imamura, K.; Ogura, T.; Kishimoto, A.; Kaminishi, M.; Esumi, H. Cell Cycle Regulation via p53 Phosphorylation by a 59-AMP Activated Protein Kinase Activator, 5-Aminoimidazole-4-Carboxamide-1-b-D-Ribofuranoside, in a Human Hepatocellular Carcinoma Cell Line. Biochem and Biophys Res Commun 2001, 287, 562–567. [Google Scholar]
- Jones, R.G.; Plas, D.R.; Kubek, S.; Buzzai, M.; Mu, J.; Xu, Y.; Birnbaum, M.J.; Thompson, C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 2005, 18, 283–293. [Google Scholar]
- Igata, M.; Motoshima, H.; Tsuruzoe, K.; Kojima, K.; Matsumura, T.; Kondo, T.; Taguchi, T.; Nakamaru, K.; Yano, M.; Kukidome, D.; Matsumoto, K.; Toyonaga, T.; Asano, T.; Nishikawa, T.; Araki, E. Adenosine monophosphate-activated protein kinase suppresses vascular smooth muscle cell proliferation through the inhibition of cell cycle progression. Circ. Res 2005, 97, 837–844. [Google Scholar]
- Mandal, S.; Guptan, P.; Owusu-Ansah, E.; Banerjee, U. Mitoch ondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell 2005, 9, 843–854. [Google Scholar]
- Owusu-Ansah, E.; Yavari, A.; Mandal, S.; Banerjee, U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet 2008, 40, 356–361. [Google Scholar]
- Portillo, F.; Mulet, J.M.; Serrano, R. A role for the non-phosphorylated form of yeast Snf1: tolerance to toxic cations and activation of potassium transport. FEBS Lett 2005, 579, 512–516. [Google Scholar]
- von Plehwe, U.; Berndt, U.; Conz, C.; Chiabudini, M.; Fitzke, E.; Sickmann, A.; Petersen, A.; Pfeifer, D.; Rospert, S. The Hsp70 Homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev 2009, 23, 2102–2115. [Google Scholar]
- Humston, E.M.; Dombek, K.M.; Hoggard, J.C.; Young, E.T.; Synovec, R.E. Time-dependent profiling of metabolites from Snf1 mutant and wild type yeast cells. Anal. Chem 2008, 80, 8002–8011. [Google Scholar]
- Estruch, F.; Treitel, M.A.; Yang, X.; Carlson, M. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 1992, 132, 639–650. [Google Scholar]
- Pessina, S.; Tsiarentsyeva, V.; Busnelli, S.; Vanoni, M.; Alberghina, L.; Coccetti, P. Snf1/AMPKpromotes S-phase entrance by controlling CLB5 transcription in budding yeast. Cell Cycle 2010, 9, 2189–2200. [Google Scholar]
- Tanaka, S.; Tak, Y.S.; Araki, H. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div 2007, 5, 2–16. [Google Scholar]
- Brümmer, A.; Salazar, C.; Zinzalla, V.; Alberghina, L.; Höfer, T. Mathematical modelling of DNA replication reveals a trade-off between coherence of origin activation and robustness against rereplication. PLoS Comp. Biol 2010, 6, e1000783. [Google Scholar]
- Liu, Y.; Xu, X.; Kuo, M.H. Snf1p regulates gcn5p transcriptional activity by antagonizing spt3p. Genetics 2010, 184, 91–105. [Google Scholar]
- Huang, D.; Kaluarachchi, S.; van Dyk, D.; Friesen, H.; Sopko, R.; Ye, W.; Bastajian, N.; Moffat, J.; Sassi, H.; Costanzo, M.; Andrews, B.J. Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast. PLoS Biol 2009, 7, e1000188. [Google Scholar]
- Wang, H.; Carey, L.B.; Cai, Y.; Wijnen, H.; Futcher, B. Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoS Biol 2009, 7, e1000189. [Google Scholar]
- Takahata, S.; Yu, Y.; Stillman, D.J. The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J 2009, 28, 3378–3389. [Google Scholar]
- Cantó, C.; Gerhart-Hines, Z.; Feige, J.N.; Lagouge, M.; Noriega, L.; Milne, J.C.; Elliott, P.J.; Puigserver, P.; Auwerx, J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009, 458, 1056–1060. [Google Scholar]
- Gadura, N.; Michels, C.A. Sequences in the N-terminal cytoplasmic domain of Saccharomyces cerevisiae maltose permease are required for vacuolar degradation but not glucose-induced internalization. Curr. Genet 2006, 50, 101–114. [Google Scholar]
- Gadura, N.; Robinson, L.C.; Michels, C.A. Glc7-Reg1 phosphatase signals to Yck1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics 2006, 172, 1427–1439. [Google Scholar]
- Chaves, R.S.; Herrero, P.; Moreno, F. Med8, a subunit of the mediator CTD complex of RNA polymerase II, directly binds to regulatory elements of SUC2 and HXK2 genes. Biochem. Biophys. Res. Commun 1999, 254, 345–350. [Google Scholar]
- Palomino, A.; Herrero, P.; Moreno, F. Rgt1, a glucose sensing transcription factor, is required for transcriptional repression of the HXK2 gene in Saccharomyces cerevisiae. Biochem. J 2005, 388, 697–703. [Google Scholar]
- Wanke, V.; Vavassori, M.; Thevelein, J.M.; Tortora, P.; Vanoni, M. Regulation of maltose utilization in Saccharomyces cerevisiae by genes of the RAS/protein kinase A pathway. FEBS Lett 1997, 402, 251–5. [Google Scholar]
- Vanoni, M.; Vai, M.; Popolo, L.; Alberghina, L. Structural heterogeneity in populations of the budding yeast Saccharomyces cerevisiae. J. Bacteriol 1983, 156, 1282–1291. [Google Scholar]
- Lord, P.G.; Wheals, A.E. Rate of cell cycle initiation of yeast cells when cell size is not a rate-determining factor. J. Cell Sci 1983, 59, 183–201. [Google Scholar]
- Brauer, M.J.; Huttenhower, C.; Airoldi, E.M.; Rosenstein, R.; Matese, J.C.; Gresham, D.; Boer, V.M.; Troyanskaya, O.G.; Botstein, D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell 2008, 19, 352–367. [Google Scholar]
- Lu, C.; Brauer, M.J.; Botstein, D. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol. Biol. Cell 2009, 20, 891–903. [Google Scholar]
- Bradley, P.H.; Brauer, M.J.; Rabinowitz, J.D.; Troyanskaya, O.G. Coordinated concentration changes of transcripts and metabolites in Saccharomyces cerevisiae. PLoS Comput. Biol 2009, 5, e1000270. [Google Scholar]
- Castrillo, J.I.; Zeef, L.A.; Hoyle, D.C.; Zhang, N.; Hayes, A.; Gardner, D.C.; Cornell, M.J.; Petty, J.; Hakes, L.; Wardleworth, L.; Rash, B.; Brown, M.; Dunn, W.B.; Broadhurst, D.; O’Donoghue, K.; Hester, S.S.; Dunkley, T.P.; Hart, S.R.; Swainston, N.; Li, P.; Gaskell, S.J.; Paton, N.W.; Lilley, K.S.; Kell, D.B.; Oliver, S.G. Growth control of the eukaryote cell: a systems biology study in yeast. J Biol 2007, 6, 4. [Google Scholar]
- Levy, S.; Ihmels, J.; Carmi, M.; Weinberger, A.; Friedlander, G.; Barkai, N. Strategy of transcription regulation in the budding yeast. PLoS One 2007, 2, 250. [Google Scholar]
- Youk, H.; van Oudenaarden, A. Growth landscape formed by perception and import of glucose in yeast. Nature 2009, 462, 875–879. [Google Scholar]
- Bhalla, U.S.; Iyengar, R. Emergent properties of networks of biological signaling pathways. Science 1999, 283, 381–387. [Google Scholar]
- Thevelein, J.M. Signal transduction in yeast. Yeast 1994, 10, 1753–1790. [Google Scholar]
- Markwardt, D.D.; Garrett, J.M.; Eberhardy, S.; Heideman, W. Activation of the Ras/cyclic AMP pathway in the yeast Saccharomyces cerevisiae does not prevent G1arrest in response to nitrogen starvation. J. Bacteriol 1995, 177, 6761–6765. [Google Scholar]
- Martin, D.E.; Hall, M.N. The expanding TOR signaling network. Curr. Opin. Cell Biol 2005, 17, 158–166. [Google Scholar]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124(3), 471–484. [Google Scholar]
- Martin, D.E.; Powers, T.; Hall, M.N. Regulation of ribosome biogenesis: where is TOR? Cell Metab 2006, 4, 259–260. [Google Scholar]
- De Virgilio, C.; Loewith, R. The TOR signalling network from yeast to man. Int. J. Biochem. Cell Biol 2006, 38, 1476–1481. [Google Scholar]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar]
- Wedaman, K.P.; Reinke, A.; Anderson, S.; Yates, J.; McCaffery, J.M.; Powers, T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 1204–1220. [Google Scholar]
- Düvel, K; Santhanam, A.; Garrett, S.; Schneper, L.; Broach, J.R. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 2003, 11, 1467–1478. [Google Scholar]
- Dechant, R.; Peter, M. Nutrient signals driving cell growth. Curr. Opin. Cell Biol 2008, 20, 678–687. [Google Scholar]
- Inoki, K.; Guan, K.L. Complexity of the TOR signaling network. Trends Cell Biol 2006, 16, 206–212. [Google Scholar]
- Barbet, N.C.; Schneider, U.; Helliwell, S.B.; Stansfield, I.; Tuite, M.F.; Hall, M.N. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 1996, 7, 25–42. [Google Scholar]
- Zinzalla, V.; Graziola, M.; Mastriani, A.; Vanoni, M.; Alberghina, L. Rapamycin-mediated G1 arrest involves regulation of the Cdk inhibitor Sic1 in Saccharomyces cerevisiae. Mol. Microbiol 2007, 63, 1482–1494. [Google Scholar]
- Shamji, A.F.; Kuruvilla, F.G.; Schreiber, S.L. Partitioning the transcriptional program induced by rapmycin among the effectors of the Tor proteins. Curr. Biol 2000, 10, 1574–1581. [Google Scholar]
- Komeili, A.; Wedaman, K.P.; O’Shea, E.K.; Powers, T. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J Cell Biol 2000, 151, 863–878. [Google Scholar]
- Dilova, I.; Aronova, S.; Chen, J.C.; Powers, T. Tor signaling and nutrient-based signals converge on Mks1p phosphorylation to regulate expression of Rtg1.Rtg3p-dependent target genes. J. Biol. Chem 2004, 279, 46527–46535. [Google Scholar]
- Tate, J.J.; Cox, K.H.; Rai, R.; Cooper, T.G. Mks1p is required for negative regulation of retrograde gene expression in Saccharomyces cerevisiae but does not affect nitrogen catabolite repression-sensitive gene expression. J. Biol. Chem 2002, 277, 20477–20482. [Google Scholar]
- Jiang, Y.; Broach, J.R. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J 1999, 18, 2782–2792. [Google Scholar]
- Düvel, K; Broach, JR. The role of phosphatases in TOR signaling in yeast. Curr Top Microbiol Immunol 2004, 279, 19–38. [Google Scholar]
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; Broach, J.R.; De Virgilio, C.; Hall, M.N.; Loewith, R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar]
- Li, H.; Tsang, C.K.; Watkins, M.; Bertram, P.G.; Zheng, X.F. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 2006, 442, 1058–1061. [Google Scholar]
- Aronova, S.; Wedaman, K.; Anderson, S.; Yates, J.; Powers, T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 2779–2794. [Google Scholar]
- Sturgill, T.W.; Cohen, A.; Diefenbacher, M.; Trautwein, M.; Martin, D.E.; Hall, M.N. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell 2008, 7, 1819–1830. [Google Scholar]
- Rohde, J.R.; Bastidas, R.; Puria, R.; Cardenas, M.E. Nutritional control via Tor signaling in Saccharomyces cerevisiae. Curr Opin. Microbiol 2008, 11, 153–160. [Google Scholar]
- Lempiäinen, H.; Uotila, A.; Urban, J.; Dohnal, I.; Ammerer, G.; Loewith, R.; Shore, D. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell 2009, 33, 704–716. [Google Scholar]
- Schawalder, S.B.; Kabani, M.; Howald, I.; Choudhury, U.; Werner, M.; Shore, D. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 2004, 432, 1058–10614. [Google Scholar]
- Wade, J.T.; Hall, D.B.; Struhl, K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 2004, 432, 1054–1058. [Google Scholar]
- Rudra, D.; Zhao, Y.; Warner, J.R. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J 2005, 24, 533–542. [Google Scholar]
- Mai, B.; Breeden, L. Xp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol. Cell Biol 1997, 17, 6491–501. [Google Scholar]
- Mai, B.; Breeden, L.L. Identification of target genes of a yeast transcriptional repressor. Methods Mol. Biol 2006, 317, 267–277. [Google Scholar]
- Ubersax, J.A.; Woodbury, E.L.; Quang, P.N.; Paraz, M.; Blethrow, J.D.; Shah., K.; Shokat, K.M.; Morgan, D.O. Targets of the cyclin-dependent kinase Cdk1. Nature 2003, 425, 859–864. [Google Scholar]
- Jorgensen, P.; Tyers, M. How cells coordinate growth and division. Curr Biol 2004, 14, R1014–1027. [Google Scholar]
- Pringle, J.R.; Hartwell, L.H. The Saccharomyces cerevisiae cell cycle. In The Molecular Biology of the Yeast Saccharomyces cerevisiae: Life Cycle and Inheritance; Strathern, J.N., Jones, E.W., Broach, J.R., Eds.; Cold Spring Harbor Laboratory, Cold Spring Harbor: New York, NY, USA, 1981; pp. 97–142. [Google Scholar]
- Bloom, J.; Cross, F.R. Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol 2007, 8, 149–160. [Google Scholar]
- Porro, D.; Vai, M.; Vanoni, M.; Alberghino, L.; Hatzis, C. Analysis and modeling of growing budding yeast populations at the single cell level. Cytometry 2009, 75, 114–120. [Google Scholar]
- Mitchison, J.M. The Biology of the Cell Cycle; Cambridge University Press: Cambridge, UK, 1971. [Google Scholar]
- Hartwell, L.H.; Unger, M.W. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J. Cell Biol 1977, 75, 422–435. [Google Scholar]
- Johnston, G.C.; Ehrhardt, C.W.; Lorincz, A.; Carter, B.L. Regulation of cell size in the yeast Saccharomyces cerevisiae. J. Bacteriol 1979, 137, 1–5. [Google Scholar]
- Porro, D.; Brambilla, L.; Alberghina, L. Glucose metabolism and cell size in continuous cultures of Saccharomyces cerevisiae. FEMS Microbiol. Lett 2003, 229, 165–171. [Google Scholar]
- Alberghina, L.; Coccetti, P.; Orlandi, I. Systems biology of the cell cycle of Saccharomyces cerevisiae: From network mining to system-level properties. Biotechnol. Adv 2009, 27, 960–978. [Google Scholar]
- Vanoni, M.; Rossi, R.L.; Querin, L.; Zinzalla, V.; Alberghina, L. Glucose modulation of cell size in yeast. Biochem. Soc. Trans 2005, 33, 294–296. [Google Scholar]
- Barberis, M.; Klipp, E.; Vanoni, M.; Alberghina, L. Cell size at S phase initiation: an emergent property of the G1/S network. PLoS Comput. Biol 2007, 3, e64. [Google Scholar]
- Mendenhall, M.D.; Hodge, A.E. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev 1998, 62, 1191–1243. [Google Scholar]
- Rupes, I. Checking cell size in yeast. Trends Genet 2002, 18, 479–485. [Google Scholar]
- Di, T.S.; Skotheim, J.M.; Bean, J.M.; Siggia, E.D.; Cross, F.R. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 2007, 448, 947–951. [Google Scholar]
- Drebot, M.A.; Barnes, C.A.; Singer, R.A.; Johnston, G.C. Genetic assessment of stationary phase for cells of the yeast Saccharomyces cerevisiae. J. Bacteriol 1990, 172, 3584–3589. [Google Scholar]
- Anghileri, P.; Branduardi, P.; Sternieri, F.; Monti, P.; Visintin, R.; Bevilacqua, A.; Alberghina, L.; Martegani, E.; Baroni, M.D. Chromosome separation and exit from mitosis in budding yeast: dependence on growth revealed by cAMP-mediated inhibition. Exp. Cell Res 1999, 250, 510–523. [Google Scholar]
- Schneper, L.A.; Krauss, R.; Miyamoto, S.; Fang, S.; Broach, J.R. The Ras/protein kinase A pathway acts in parallel with the Mob2/Cbk1 pathway to effect cell cycle progression and proper bud site selection. Eukaryot. Cell 2004, 3, 108–120. [Google Scholar]
- Baroni, M.D.; Martegani, E.; Monti, P.; Alberghina, L. Cell size modulation by CDC25 and RAS2 genes in Saccharomyces cerevisiae. Mol. Cell. Biol 1989, 9, 2715–2723. [Google Scholar]
- Mitsuzawa, H. Increases in cell size at START caused by hyperactivation of the cAMP pathway in Saccharomyces cerevisiae. Mol. Gen. Genet 1994, 243, 158–165. [Google Scholar]
- Baroni, M.D.; Monti, P.; Marconi, G.; Alberghina, L. cAMP-mediated increase in the critical cell size required for the G1 to S transition in Saccharomyces cerevisiae. Exp. Cell Res 1992, 201, 299–306. [Google Scholar]
- Baroni, M.D.; Monti, P.; Alberghina, L. Repression of growth-regulated G1 cyclin expression by cyclic AMP in budding yeast. Nature 1994, 371, 339–342. [Google Scholar]
- Tokiwa, G.; Tyers, M.; Volpe, T.; Futcher, B. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature 1994, 371, 342–345. [Google Scholar]
- Belotti, F.; Tisi, R.; Martegani, E. The N-terminal region of the Saccharomyces cerevisiae RasGEF Cdc25 is required for nutrient dependent cell-size regulation. Microbiology 2006, 152, 1231–1242. [Google Scholar]
- Cameron, S.; Levin, L.; Zoller, M.; Wigler, M. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell 1988, 53, 555–566. [Google Scholar]
- Jorgensen, P.; Nishikawa, J.L.; Breitkreutz, B.J.; Tyers, M. Systematic identification of pathways that couple cell growth and division in yeast. Science 2002, 297, 395–400. [Google Scholar]
- Levy, S; Barkai, N. Coordination of gene expression with growth rate: a feedback or a feed-forward strategy? FEBS Lett 2009, 583, 3974–3978. [Google Scholar]



© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Busti, S.; Coccetti, P.; Alberghina, L.; Vanoni, M. Glucose Signaling-Mediated Coordination of Cell Growth and Cell Cycle in Saccharomyces Cerevisiae. Sensors 2010, 10, 6195-6240. https://doi.org/10.3390/s100606195
Busti S, Coccetti P, Alberghina L, Vanoni M. Glucose Signaling-Mediated Coordination of Cell Growth and Cell Cycle in Saccharomyces Cerevisiae. Sensors. 2010; 10(6):6195-6240. https://doi.org/10.3390/s100606195
Chicago/Turabian StyleBusti, Stefano, Paola Coccetti, Lilia Alberghina, and Marco Vanoni. 2010. "Glucose Signaling-Mediated Coordination of Cell Growth and Cell Cycle in Saccharomyces Cerevisiae" Sensors 10, no. 6: 6195-6240. https://doi.org/10.3390/s100606195
APA StyleBusti, S., Coccetti, P., Alberghina, L., & Vanoni, M. (2010). Glucose Signaling-Mediated Coordination of Cell Growth and Cell Cycle in Saccharomyces Cerevisiae. Sensors, 10(6), 6195-6240. https://doi.org/10.3390/s100606195




