Synthesis and Characterization of Polyaniline/Graphene Composite Nanofiber and Its Application as an Electrochemical DNA Biosensor for the Detection of Mycobacterium tuberculosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
- Probe DNA-single stranded DNA (ssDNA):5′-CTC gTC CAg CgC CgC TTC gg-3′
- Complementary target DNA:5′-CCg AAg Cgg CgC Tgg ACg Ag-3′
- Non-complementary DNA:5′-TTT GGT ATT ATT GTT CAT GT-3′.
2.2. Synthesis of PANI and PANI/GP Nanofiber
2.3. Fabrication of DNA Biosensor
2.4. Electrochemical Measurements
2.5. Characterization
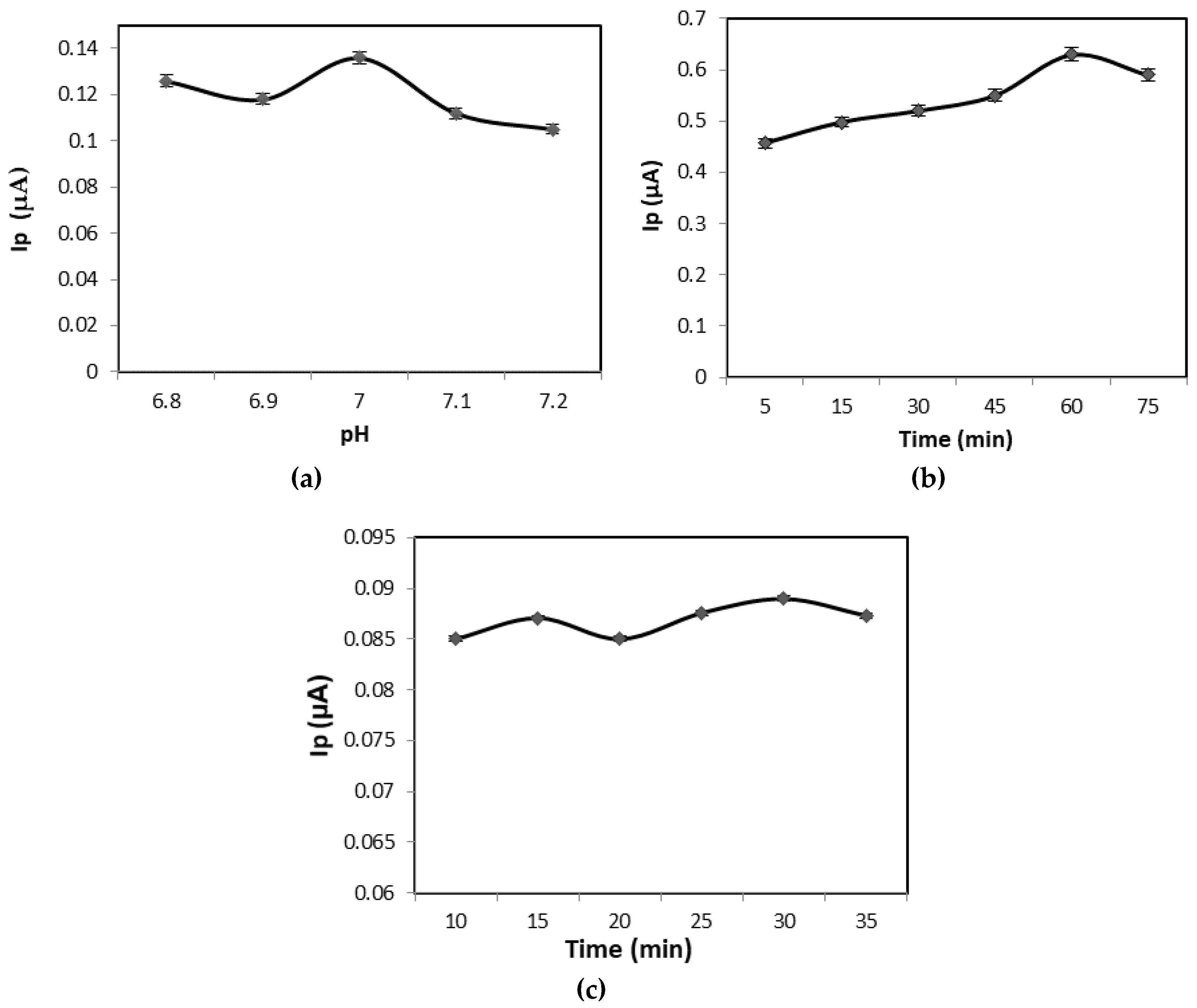
3. Results and Discussion
Electrochemical Characterization of the DNA Biosensor
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Arentz, M.; Hawn, T.R. Tuberculosis Infection: Insight from Immunogenomics. Drug Discov. Today Dis. Mech. 2007, 4, 231–236. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Global Tuberculosis Report 2012. Available online: www.who.int/iris/bitstream/10665/75938/1/9789241564502_eng.pdf (accessed on 25 June 2017).
- Thiruppathiraja, C.; Kamatchiammal, S.; Adaikkappan, P.; Santhosh, D.J.; Alagar, M. Specific detection of Mycobacterium sp. genomic DNA using dual labeled gold nanoparticle based electrochemical biosensor. Anal. Biochem. 2011, 417, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Garberi, J.; Labrador, J.; Garberi, F.; Garberi, J.E.; Peneipil, J.; Garberi, M.; Scigliano, L.; Troncoso, A. Diagnosis of Mycobacterium tuberculosis using molecular biology technology. Asian Pac. J. Trop. Biomed. 2011, 1, 89–93. [Google Scholar] [CrossRef]
- Soo, P.C.; Horng, Y.T.; Chang, C.; Wang, J.Y.; Hsueh, P.R.; Chuang, C.Y.; Lu, C.C.; Lai, H.C. A simple gold nanoparticle probes assay for identification of Mycobacterium tuberculosis and Mycobacterium tuberculosis complex from clinical specimens. Mol. Cell. Probes 2009, 23, 240–246. [Google Scholar] [CrossRef] [PubMed]
- The Malay Mail online. TB Cases Increased by 6pc in 2016, Says Health Minister. Available online: http://www.themalaymailonline.com/malaysia/article/tb-cases-increased-by-6pc-in-2016-says-health-minister#THzWfPWlY12eS2rg.97 (accessed on 30 October 2017).
- Fukunaga, H.; Murukami, T.; Gondo, T.; Sugi, K.; Ishihara, T. Sensitivity of acid-fast staining for Mycobacterium tuberculosis in formalin-fixed tissue. Am. J. Respir. Crit. Care Med. 2002, 166, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.R.; Shenai, S.; Desai, D.C. Comparison of Mycobacterium tuberculosis culture using liquid culture medium and Lowenstein Jensen medium in abdominal tuberculosis. Indian J. Gastroenterol. 2010, 29, 237–239. [Google Scholar] [CrossRef] [PubMed]
- Bardarov, S., Jr.; Dou, S.; Eisenach, K.; Banaiee, N.; Ya, S.; Chan, J. Detection and drug-susceptibility testing of M. tuberculosis from sputum samples using luciferase reporter phage: Comparison with the Mycobacteria Growth Indicator Tube (MGIT) system. Diag. Microbiol. Infect. Dis. 2003, 45, 53–61. [Google Scholar] [CrossRef]
- Li, H.; Turhan, V.; Chokhani, L.; Stratton, C.W.; Dunbar, S.A.; Tang, Y.W. Identification and differentiation of clinically relevant mycobacterium species directly from acid-fast bacillus-positive culture broth. Clin. Microbiol. 2009, 47, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.F.; Curry, J.I. Detection and identification of mycobacterium tuberculosis directly from sputum sediments by ligase chain reaction. J. Clin. Microbiol. 1998, 36, 1028–1031. [Google Scholar] [PubMed]
- Hsieh, S.C.; Chang, C.C.; Lu, C.C.; Wei, C.F.; Lin, C.S.; Lai, H.-C.; Lin, C.-W. Rapid identification of Mycobacterium tuberculosis infection by a new array format-based surface plasmon resonance method. Nanoscale Res. Lett. 2012, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yan, W.; Vijapur, D.; Botte, G.G. Electrochemically reduced graphene oxide—Nickel nanocomposites for urea electrolysis. Electrochim. Acta 2013, 89, 732–736. [Google Scholar] [CrossRef]
- Liu, C.; Jang, D.; Xiang, G.; Liu, F.; Liu, L.; Pu, X. An electrochemical DNA biosensor for sensitive Mycobacterium tuberculosis detection based on signal amplification of graphene and gold nanoparticles-polyaniline nanocomposite. Analyst 2014, 10, 5460–5465. [Google Scholar] [CrossRef] [PubMed]
- Kaewphinit, T.; Santiwatanakul, S.; Promptmas, C.; Chansiri, K. Detection of non-amplified Mycobacterium tuberculosis genomic DNA using piezoelectric DNA-based biosensors. Sensors 2010, 10, 1846–1858. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, N.; Arora, K.; Arya, S.K.; Solanki, P.R.; Iwamoto, M.; Malhotra, B.D. Nucleic acid sensor for M. tuberculosis detection based on surface plasmon resonance. Analyst 2008, 1, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Pang, P.; Cai, Q.; Yao, S.; Grimes, C.A. The detection of Mycobacterium tuberculosis in sputum sample based on a wireless magnetoelastic sensing device. Talanta 2008, 76, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Abu-salah, K.M.; Zourob, M.M.; Mouffouk, F.; Alrokayan, S.A.; Alaamery, M.A.; Ansari, A.A. DNA-based nano biosensors as an emerging platform for detection of disease. Sensors 2015, 15, 14539–14568. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; He, X.; He, D.; Wang, K.; Qin, K. Biosensing technologies for mycobacterium tuberculosis detection: Status and new developments. Clin. Dev. Immunol. 2011, 2011, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Sumana, G.; Nagarajan, R.; Malhotra, B.D. Zirconia based nucleic acid sensor for Mycobacterium tuberculosis detection. Appl. Phys. Lett. 2010, 96, 1050–1058. [Google Scholar] [CrossRef] [Green Version]
- Arora, K.; Prabhakar, N.; Chand, S.; Malhotra, B.D. Escherichia coli genosensor based on polyaniline. Anal. Chem. 2007, 79, 6152–6158. [Google Scholar] [CrossRef] [PubMed]
- Dhand, C.; Das, M.; Datta, M.; Malhotra, B.D. Recent advances in polyaniline based biosensors. Biosens. Bioelectron. 2011, 26, 2811–2821. [Google Scholar] [CrossRef] [PubMed]
- Montenegro, S.H.; Gilman, R.H.; Sheen, P.; Cama, R.; Caviedes, L.; Hopper, T.; Chambers, R.; Oberhelman, R.A. Improved detection of Mycobacterium tuberculosis in Peruvian children by use of a heminested IS6110 polymerase chain reaction assay. Clin. Infect. Dis. 2003, 36, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.R.; Munichandraiah, N. Fabrication and evaluation of 450 F electrochemical redox supercapacitors using inexpensive and high-performance, polyaniline coated, stainless-steel electrodes. J. Power Sources 2002, 112, 443–451. [Google Scholar] [CrossRef]
- Jiang, L.; Yuan, R.; Bai, L.; Yuan, Y.; Chai, Y.; Wang, Y. Aptamer-based highly sensitive electrochemical detection of thrombin via the amplification of graphene. Analyst 2012, 137, 2415–2420. [Google Scholar] [CrossRef] [PubMed]
- Geim, A.K.; Novoselov, K.S. The rise of graphene. Nat. Mater. 2007, 6, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Murali, S.; Cai, W.; Li, X.; Suk, J.W.; Potts, J.R.; Ruoff, R.S. Graphene and graphene oxide: Synthesis, properties, and applications. Adv. Mater. 2010, 22, 3906–3924. [Google Scholar] [CrossRef] [PubMed]
- Torati, S.R.; Reddy, V.; Yoon, S.S.; Kim, C. Electrochemical biosensor for Mycobacterium tuberculosis DNA detection based on gold nanotubes array electrode platform. Biosens. Bioelectron. 2015, 78, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hao, Q.; Yang, X.; Lu, L.; Wang, X. Graphene oxide doped-polyaniline for supercapacitors. Electrochem. Commun. 2009, 11, 1158–1161. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, H.; Hsu, C.F.; Kilmartin, P.A.; Travas-Sejdic, J. Self-assembled polyaniline nanotubes grown from a polymeric acid solution. Nanotechnology 2007, 18, 607–614. [Google Scholar] [CrossRef]
- Zhang, Z.; Wei, Z.; Wan, M. Nanostructures of polyaniline doped with inorganic acids. Macromolecules 2002, 35, 5937–5942. [Google Scholar] [CrossRef]
- Venancio, E.C.; Wang, P.-C.; MacDiarmid, A.G. The azanes: A class of material incorporating nano/microself-assembled hollow spheres obtained by aqueous oxidative polymerization of aniline. Synth. Met. 2006, 156, 357–369. [Google Scholar] [CrossRef]
- Trchova, M.; Sedenkova, I.; Konyushenko, E.N.; Stejskal, J.; Holler, P.; Ciric-Marjanovic, G. Evolution of polyaniline nanotubes: The oxidation of aniline in water. J. Phys. Chem. B 2006, 110, 9461–9468. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Cho, M.S.; Choi, H.J.; Jhon, M.S. Synthesis of polyaniline using stabilizer and its electrorheological properties. Synth. Met. 2003, 135, 21–22. [Google Scholar] [CrossRef]
- Koo, C.M.; Jeon, B.H.; Chung, I.J. The effect of poly(methyl vinyl ether-alt-maleic acid) stabilizer on the stability of polyaniline-poly(methyl vinyl ether-alt-maleic acid) dispersions. J. Colloid Interface Sci. 2000, 227, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wan, M. Synthesis and characterization of self-assembled polyaniline nanotubes doped with D-10-camphorsulfonic acid. Nanotechnology 2002, 13, 750–755. [Google Scholar] [CrossRef]
- Kim, B.J.; Oh, S.G.; Han, M.G.; Im, S.S. Preparation of polyaniline nanoparticles in micellar solutions as polymerization medium. Langmuir 2000, 16, 5841–5845. [Google Scholar] [CrossRef]
- Harada, M.; Adachi, M. Surfactant-mediated fabrication of silica nanotubes. Adv. Mater. 2000, 12, 839–841. [Google Scholar] [CrossRef]
- Wang, L.; Yao, Q.; Bi, H.; Huang, F.; Wang, Q.; Chen, L. PANI/graphene nanocomposite films with high thermoelectric properties by enhanced molecular ordering. J. Mater. Chem. A 2015, 3, 7086–7092. [Google Scholar] [CrossRef]
- Chen, S.A.; Lee, H.T. Structure and properties of poly(acrylic acid)-doped polyaniline. Macromolecules 1995, 28, 2858–2866. [Google Scholar] [CrossRef]
- Furukawa, Y.; Ueda, F.; Hyodo, Y.; Harada, I.; Nakajima, F.; Kawagoe, T. Vibrational spectra and structure of polyaniline. Macromolecules 1998, 21, 1297–1305. [Google Scholar] [CrossRef]
- Sariciftci, N.S.; Kuzmany, H.; Neugebauer, H.; Neckel, A. Structural and electronic transitions in polyaniline: A Fourier transforms infrared spectroscopic study. J. Chem. Phys. 1990, 92, 4530–4539. [Google Scholar] [CrossRef]
- Sapurina, I.; Mokeev, M.; Lavrentev, V.; Zgonnik, V.; Trchova, M.; Hlavata, D.; Stejskal, J. Polyaniline complex with fullerene C60. Eur. Polym. J. 2000, 36, 2321–2326. [Google Scholar] [CrossRef]
- Khutoryanskaya, O.V.; Khutoryanskiy, V.; Pethrick, R.A. Characterisation of blends based on hydroxyethylcellulose and maleic acid-alt-methyl vinyl ether. Macromol. Chem. Phys. 2005, 206, 1497–1510. [Google Scholar] [CrossRef]
- Wang, L.; Bi, H.; Yao, Q.; Ren, D.; Qu, S.; Huang, F.; Chen, L. Three-dimensional tubular graphene/polyaniline composites as high-performance elastic thermoelectrics. Compos. Sci. Technol. 2017, 150. [Google Scholar] [CrossRef]
- Mundinamani, S.P.; Rabinal, M.K. Cyclic voltammetric studies on the role of electrode, electrode surface modification and electrolyte solution of an electrochemical cell. J. Appl. Chem. 2014, 7, 45–52. [Google Scholar]
- Pandurangachar, M.; Swamy, B.E.K.; Chandrashekar, B.N.; Gilbert, O.; Reddy, S. Electrochemical investigations of potassium ferricyanide and dopamine by1-butyl-4-methylpyridinium tetrafluoro borate modified carbon paste electrode: A cyclic voltammetric study. Int. J. Electrochem. Sci. 2010, 5, 1187–1202. [Google Scholar]
- Sung, M.S.; Hwang, W. Amperometric hydrogen peroxide biosensor based on a modified gold electrode with silver nanowires. J. Appl. Electrochem. 2010, 1, 279–285. [Google Scholar] [CrossRef]
- Konopka, S.J.; Mcduffie, B. Diffusion coefficients of ferri- and ferrocyanide ions in aqueous media, using twin-electrode thin-layer electrochemistry. Anal. Chem. 1970, 42, 1741–1746. [Google Scholar] [CrossRef]
- Mat Zaid, M.H.; Abdullah, J.; Yusof, N.A.; Sulaiman, Y.; Wasoh, H.; Md Nor, M.F.; Issa, R. PNA biosensor based graphene oxide/water soluble quantum dots for detection of Mycobaterium tuberculosis. Sens. Actuators B Chem. 2016, 241, 1024–1034. [Google Scholar] [CrossRef]
- Zhu, L.; Zhao, R.; Wang, K.; Xiang, H.; Shang, Z.; Sun, W. Electrochemical behaviors of methylene blue on DNA modified electrode and its application to the detection of PCR product from NOS sequence. Sensors 2008, 8, 5649–5660. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.Y.; Lu, X.J.; Ju, H.X. DNA sensor for recognition of native yeast DNA sequence with methylene blue as an electrochemical hybridization indicator. Electroanalysis 2002, 14, 949–954. [Google Scholar] [CrossRef]
- Kelley, S.O.; Boon, E.M.; Barton, J.K.; Jackson, N.M.; Hill, M.G. Single-base mismatch detection based on charge transduction through DNA. Nucleic Acids Res. 1999, 27, 4830–4837. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, D.; Kara, P.; Kerman, K.; Meric, B.; Erdem, A.; Jelen, F.; Nielsen, P.E.; Ozsoz, M. DNA and PNA sensing on mercury and carbon electrodes by using methylene blue as an electrochemical label. Bioelectrochemistry 2002, 58, 119–126. [Google Scholar] [CrossRef]
- Sun, H.; Wu, L.; Wei, W.; Qu, X. Recent advances in graphene quantum dots for sensing. Mater. Today 2013, 16, 433–442. [Google Scholar] [CrossRef]
- Kerman, K.; Ozkan, D.; Kara, P.; Meric, B.; Gooding, J.J.; Ozsoz, M. Voltammetric determination of DNA hybridization using methylene blue and self-assembled alkane thiol monolayer on gold electrodes. Anal. Chim. Acta 2002, 462, 39–47. [Google Scholar] [CrossRef]
- Pan, D.; Zuo, X.; Wan, Y.; Wang, L.; Zhang, L.; Song, S.; Fan, C. Electrochemical interrogation of interactions between surface-confined DNA and methylene blue. Sensors 2007, 7, 2671–2680. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.N.; Miller, J.C. Statistics and Chemometrics for Analytical Chemistry, 6th ed.; Pearson Education: North York, ON, Canada, 2010; pp. 153–154. [Google Scholar]












© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamad, F.S.; Mat Zaid, M.H.; Abdullah, J.; Zawawi, R.M.; Lim, H.N.; Sulaiman, Y.; Abdul Rahman, N. Synthesis and Characterization of Polyaniline/Graphene Composite Nanofiber and Its Application as an Electrochemical DNA Biosensor for the Detection of Mycobacterium tuberculosis. Sensors 2017, 17, 2789. https://doi.org/10.3390/s17122789
Mohamad FS, Mat Zaid MH, Abdullah J, Zawawi RM, Lim HN, Sulaiman Y, Abdul Rahman N. Synthesis and Characterization of Polyaniline/Graphene Composite Nanofiber and Its Application as an Electrochemical DNA Biosensor for the Detection of Mycobacterium tuberculosis. Sensors. 2017; 17(12):2789. https://doi.org/10.3390/s17122789
Chicago/Turabian StyleMohamad, Fatimah Syahidah, Mohd Hazani Mat Zaid, Jaafar Abdullah, Ruzniza Mohd Zawawi, Hong Ngee Lim, Yusran Sulaiman, and Norizah Abdul Rahman. 2017. "Synthesis and Characterization of Polyaniline/Graphene Composite Nanofiber and Its Application as an Electrochemical DNA Biosensor for the Detection of Mycobacterium tuberculosis" Sensors 17, no. 12: 2789. https://doi.org/10.3390/s17122789
APA StyleMohamad, F. S., Mat Zaid, M. H., Abdullah, J., Zawawi, R. M., Lim, H. N., Sulaiman, Y., & Abdul Rahman, N. (2017). Synthesis and Characterization of Polyaniline/Graphene Composite Nanofiber and Its Application as an Electrochemical DNA Biosensor for the Detection of Mycobacterium tuberculosis. Sensors, 17(12), 2789. https://doi.org/10.3390/s17122789





