Amplified Detection of the Aptamer–Vanillin Complex with the Use of Bsm DNA Polymerase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. ISFET Fabrication
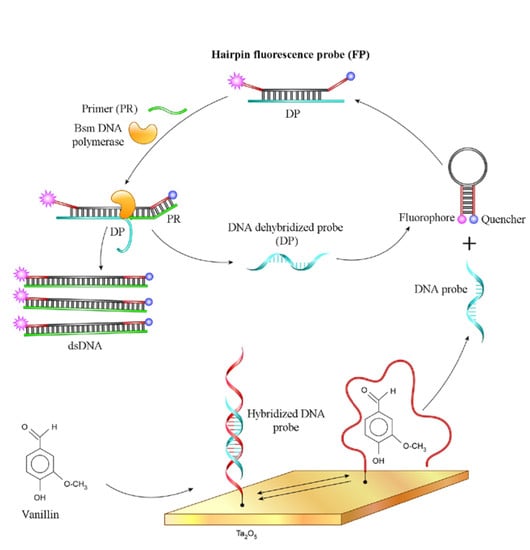
2.3. Fluorescence Measurement of Bsm DNA Polymerase Reaction
2.4. Aptamer Selection
2.5. Aptamer Immobilization
2.6. ISFET Measurements
2.7. Aptamer Characterization by PAGE [28]
3. Results
3.1. Optimization of Operating Conditions
- Aptamer for vanillin could not perform in the Bsm buffer and could bind vanillin in the Selection buffer and low molarity selection buffer.
- Bsm DNA polymerase is active in a low molarity selection buffer and the greatest activity is with DP compared to B1.
- B1 probe can be replaced by DP as a dehybridization probe from the aptamer Van_74 during the vanillin addition.
3.2. Bsm DNA Polymerase Reaction on the ISFET
3.3. Amplified Detection of the Aptamer–Vanillin Complex with the Bsm DNA Polymerase and ISFET
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rajendran, M.; Ellington, A.D. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal. Bioanal. Chem. 2008, 390, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Nikolaus, N.; Strehlitz, B. Capture-SELEX: Selection of DNA Aptamers for Aminoglycoside Antibiotics. J. Anal. Methods Chem. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, M.N.; de Prada, P.; Landry, D.W. Aptamer-Based Folding Fluorescent Sensor for Cocaine. J. Am. Chem. Soc. 2001, 123, 4928–4931. [Google Scholar] [CrossRef] [PubMed]
- Huizenga, D.E.; Szostak, J.W. A DNA Aptamer That Binds Adenosine and ATP. Biochemistry 1995, 34, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, G.R.; Jenison, R.D.; Wick, C.L.; Simorre, J.-P.; Pardi, A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat. Struct. Biol. 1997, 4, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434. [Google Scholar] [CrossRef] [PubMed]
- Ilgu, M.; Nilsen-Hamilton, M. Aptamers in analytics. Analyst 2016, 141, 1551–1568. [Google Scholar] [CrossRef] [PubMed]
- Swensen, J.S.; Xiao, Y.; Ferguson, B.S.; Lubin, A.A.; Lai, R.Y.; Heeger, A.J.; Plaxco, K.W.; Soh, H.T. Continuous, Real-Time Monitoring of Cocaine in Undiluted Blood Serum via a Microfluidic, Electrochemical Aptamer-Based Sensor. J. Am. Chem. Soc. 2009, 131, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Ferapontova, E.E.; Olsen, E.M.; Gothelf, K.V. An RNA Aptamer-Based Electrochemical Biosensor for Detection of Theophylline in Serum. J. Am. Chem. Soc. 2008, 130, 4256–4258. [Google Scholar] [CrossRef] [PubMed]
- Rowe, A.A.; Miller, E.A.; Plaxco, K.W. Reagentless Measurement of Aminoglycoside Antibiotics in Blood Serum via an Electrochemical, Ribonucleic Acid Aptamer-Based Biosensor. Anal. Chem. 2010, 82, 7090–7095. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xiao, X.; Xu, S.; Liu, Y.; Li, Y. Electrochemical aptamer-based nanosensor fabricated on single Au nanowire electrodes for adenosine triphosphate assay. Biosens. Bioelectron. 2018, 99, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ellis, A.V.; Kobus, H.; Voelcker, N.H. Aptamer sensor for cocaine using minor groove binder based energy transfer. Anal. Chim. Acta 2012, 719, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Che, X.; Que, L.; Chen, C.; Wang, W. Rapid detection of theophylline using aptamer-based nanopore thin film sensor. In Proceedings of the 2016 IEEE SENSORS, Orlando, FL, USA, 30 October–3 November 2016; pp. 1–3. [Google Scholar]
- Ramezani, M.; Mohammad Danesh, N.; Lavaee, P.; Abnous, K.; Mohammad Taghdisi, S. A novel colorimetric triple-helix molecular switch aptasensor for ultrasensitive detection of tetracycline. Biosens. Bioelectron. 2015, 70, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, V.; Xiao, Y.; Shlyahovsky, B.; Willner, I. Aptamer-Functionalized Au Nanoparticles for the Amplified Optical Detection of Thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.A.; Wang, J.; Kawde, A.-N.; Xiang, Y.; Gothelf, K.V.; Collins, G. Quantum-Dot/Aptamer-Based Ultrasensitive Multi-Analyte Electrochemical Biosensor. J. Am. Chem. Soc. 2006, 128, 2228–2229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiang, B.; Xiang, Y.; Zhang, Y.; Chai, Y.; Yuan, R. Aptamer/quantum dot-based simultaneous electrochemical detection of multiple small molecules. Anal. Chim. Acta 2011, 688, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Qi, Q.; Wang, X.; Bie, P. Porous platinum nanotubes labeled with hemin/G-quadruplex based electrochemical aptasensor for sensitive thrombin analysis via the cascade signal amplification. Biosens. Bioelectron. 2014, 57, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Piorek, B.D.; Plaxco, K.W.; Heeger, A.J. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J. Am. Chem. Soc. 2005, 127, 17990–17991. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.R.; Lai, R.Y.; Wood, M.S.; Doctor, E.H.; Heeger, A.J.; Plaxco, K.W. An Electronic, Aptamer-Based Small-Molecule Sensor for the Rapid, Label-Free Detection of Cocaine in Adulterated Samples and Biological Fluids. J. Am. Chem. Soc. 2006, 128, 3138–3139. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Morita, K.; Sato, Y.; Dai, Q.; Nishizawa, S.; Teramae, N. Label-free aptamer-based sensor using abasic site-containing DNA and a nucleobase-specific fluorescent ligand. Chem. Commun. 2009, 6445. [Google Scholar] [CrossRef] [PubMed]
- Bergveld, P. Development of an ion-sensitive solid-state device for neurophysiological measurements. IEEE Trans. Biomed. Eng. 1970, 17, 70–71. [Google Scholar] [CrossRef] [PubMed]
- Zayats, M.; Huang, Y.; Gill, R.; Ma, C.; Willner, I. Label-free and reagentless aptamer-based sensors for small molecules. J. Am. Chem. Soc. 2006, 128, 13666–13667. [Google Scholar] [CrossRef] [PubMed]
- Sharon, E.; Freeman, R.; Tel-Vered, R.; Willner, I. Impedimetric or Ion-Sensitive Field-Effect Transistor (ISFET) Aptasensors Based on the Self-Assembly of Au Nanoparticle-Functionalized Supramolecular Aptamer Nanostructures. Electroanalysis 2009, 21, 1291–1296. [Google Scholar] [CrossRef]
- Anand, A.; Liu, C.-R.; Chou, A.-C.; Hsu, W.-H.; Ulaganathan, R.K.; Lin, Y.-C.; Dai, C.-A.; Tseng, F.-G.; Pan, C.-Y.; Chen, Y.-T. Detection of K + Efflux from Stimulated Cortical Neurons by an Aptamer-Modified Silicon Nanowire Field-Effect Transistor. ACS Sens. 2017, 2, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Goda, T.; Miyahara, Y. A hairpin DNA aptamer coupled with groove binders as a smart switch for a field-effect transistor biosensor. Biosens. Bioelectron. 2012, 32, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Yang, X.; Wang, K.; Tan, W.; Li, W.; Tang, H.; Li, H. Sensitive fluorescence detection of nucleic acids based on isothermal circular strand-displacement polymerization reaction. Nucl. Acids Res. 2009, 37, e20. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, A.E.; Komarova, N.V.; Andrianova, M.S.; Grudtsov, V.P.; Kuznetsov, E.V. Aptamer based vanillin sensor using an ion-sensitive field-effect transistor. Microchim. Acta 2017. [Google Scholar] [CrossRef]
- Kuznetsov, A.; Andrianova, M.; Komarova, N.; Grudtsov, V.; Kuznetsov, E.; Saurov, A. Detection of aroma compound by ISFET modified with aptamer. In Proceedings of the 2017 2nd International Conference on Bio-engineering for Smart Technologies (BioSMART), Paris, France, 30 August–1 September 2017; pp. 1–3. [Google Scholar]
- Gubanova, O.; Andrianova, M.; Saveliev, M.; Komarova, N.; Kuznetsov, E.; Kuznetsov, A. Fabrication and package of ISFET biosensor for micro volume analysis with the use of direct ink writing approach. Mater. Sci. Semicond. Process. 2016. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl. Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- SantaLucia, J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 1998, 95, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Hianik, T.; Ostatná, V.; Sonlajtnerova, M.; Grman, I. Influence of ionic strength, pH and aptamer configuration for binding affinity to thrombin. Bioelectrochemistry 2007, 70, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bing, T.; Mei, H.; Fang, C.; Cao, Z.; Shangguan, D. Characterization and application of a DNA aptamer binding to l-tryptophan. Analyst 2011, 136, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.-H.; Yen, S.-L.; Lin, M.-S.; Chang, Y.; Louis, S.R.; Higuchi, A.; Chen, W.-Y. Microcalorimetrics Studies of the Thermodynamics and Binding Mechanism between l-Tyrosinamide and Aptamer. J. Phys. Chem. B 2008, 112, 6665–6673. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, C.; Li, X.; Song, Y.; Wang, W.; Li, C.; Hu, J.; Zhu, Z.; Li, J.; Zhang, W.; et al. Identification, Characterization and Application of a G-Quadruplex Structured DNA Aptamer against Cancer Biomarker Protein Anterior Gradient Homolog 2. PLoS ONE 2012, 7, e46393. [Google Scholar] [CrossRef] [PubMed]
- Stern, E.; Wagner, R.; Sigworth, F.J.; Breaker, R.; Fahmy, T.M.; Reed, M.A. Importance of the Debye Screening Length on Nanowire Field Effect Transistor Sensors. Nano Lett. 2007, 7, 3405–3409. [Google Scholar] [CrossRef] [PubMed]
- Bunimovich, Y.L.; Shin, Y.S.; Yeo, W.-S.; Amori, M.; Kwong, G.; Heath, J.R. Quantitative Real-Time Measurements of DNA Hybridization with Alkylated Nonoxidized Silicon Nanowires in Electrolyte Solution. J. Am. Chem. Soc. 2006, 128, 16323–16331. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y. DNA polymerases drive DNA sequencing-by-synthesis technologies: Both past and present. Front. Microbiol. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Meshik, X.; Choi, M.; Farid, S.; Datta, D.; Lan, Y.; Poduri, S.; Sarkar, K.; Baterdene, U.; Huang, C.-E.; et al. A Graphene and Aptamer Based Liquid Gated FET-Like Electrochemical Biosensor to Detect Adenosine Triphosphate. IEEE Trans. Nanobiosci. 2015, 14, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jo, M.; Kim, T.H.; Ahn, J.-Y.; Lee, D.; Kim, S.; Hong, S. Aptamer sandwich-based carbon nanotube sensors for single-carbon-atomic-resolution detection ofnon-polar small molecular species. Lab Chip 2011, 11, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Sharon, E.; Liu, X.; Freeman, R.; Yehezkeli, O.; Willner, I. Label-Free Analysis of Thrombin or Hg2+ Ions by Nucleic Acid-Functionalized Graphene Oxide Matrices Assembled on Field-Effect Transistors. Electroanalysis 2013, 25, 851–856. [Google Scholar] [CrossRef]
- Andrianova, M.S.; Grudtsov, V.P.; Komarova, N.V.; Kuznetsov, E.V.; Kuznetsov, A.E. ISFET-based Aptasensor for Thrombin Detection Using Horseradish Peroxidase. Procedia Eng. 2017, 174, 1084–1092. [Google Scholar] [CrossRef]
- Yoon, H.; Kim, J.-H.; Lee, N.; Kim, B.-G.; Jang, J. A Novel Sensor Platform Based on Aptamer-Conjugated Polypyrrole Nanotubes for Label-Free Electrochemical Protein Detection. ChemBioChem 2008, 9, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Pacios, M.; Martin-Fernandez, I.; Borrisé, X.; del Valle, M.; Bartrolí, J.; Lora-Tamayo, E.; Godignon, P.; Pérez-Murano, F.; Esplandiu, M.J. Real time protein recognition in a liquid-gated carbon nanotube field-effect transistor modified with aptamers. Nanoscale 2012, 4, 5917. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Kim, K.S.; Kim, C.-J.; Hahn, S.K.; Jo, M.-H. Electrical detection of VEGFs for cancer diagnoses using anti-vascular endotherial growth factor aptamer-modified Si nanowire FETs. Biosens. Bioelectron. 2009, 24, 1801–1805. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Maehashi, K.; Matsumoto, K. Label-Free Biosensors Based on Aptamer-Modified Graphene Field-Effect Transistors. J. Am. Chem. Soc. 2010, 132, 18012–18013. [Google Scholar] [CrossRef] [PubMed]
- Maehashi, K.; Katsura, T.; Kerman, K.; Takamura, Y.; Matsumoto, K.; Tamiya, E. Label-Free Protein Biosensor Based on Aptamer-Modified Carbon Nanotube Field-Effect Transistors. Anal. Chem. 2007, 79, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Farid, S.; Meshik, X.; Choi, M.; Mukherjee, S.; Lan, Y.; Parikh, D.; Poduri, S.; Baterdene, U.; Huang, C.-E.; Wang, Y.Y.; et al. Detection of Interferon gamma using graphene and aptamer based FET-like electrochemical biosensor. Biosens. Bioelectron. 2015, 71, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.; Lee, J.S.; Shin, D.H.; Jang, J. Aptamer-Functionalized Hybrid Carbon Nanofiber FET-Type Electrode for a Highly Sensitive and Selective Platelet-Derived Growth Factor Biosensor. ACS Appl. Mater. Interfaces 2014, 6, 13859–13865. [Google Scholar] [CrossRef] [PubMed]
- Ruslinda, A.R.; Tajima, S.; Ishii, Y.; Ishiyama, Y.; Edgington, R.; Kawarada, H. Aptamer-based biosensor for sensitive PDGF detection using diamond transistor. Biosens. Bioelectron. 2010, 26, 1599–1604. [Google Scholar] [CrossRef] [PubMed]






| Sequence Name | Sequence, 5′-3′ Direction | Comment |
|---|---|---|
| Van_74 | CGACCAGCTCATTCCTCAGGAGAAACATGGAGTCTCGATGATAGTAGGAGCGGCGGA ACGTAGGAAGAGAGGATGACGGAGGATCCGAGCTCACCAGTC | Aptamer for vanillin |
| B1 | CATCGAGACTCC | Capture probe without biotin |
| DP | ACCACATCGAGACTCCTGTGTCCTTT | Bsm dehybridization probe |
| FP | FAM-TCTTGGFCFCAGGAGTCTCGATGTGGTATTGTGTCCAAGA-BHQ1 | Fluorescence probe |
| PR | TCTTGGAC | Primer |
| Substance | LoD, M | Type | Reference |
|---|---|---|---|
| Low-weight molecules | |||
| Adenosine | 5 × 10−5 | Si-ISFET | [23] |
| Adenosine | 1 × 10−11 | Graphene-FET | [40] |
| Cocaine | 1 × 10−6 | Si-ISFET | [24] |
| K+ | Kass = (2.18 ± 0.44) × 106 | Si-ISFET | [25] |
| Bisphenol A | 1 × 10−12–1 × 10−14 | Carbon-FET | [41] |
| ATP | - | FET | [26] |
| Vanillin (our previous work) | 1.55 × 10−7 | Si-ISFET | [28] |
| Vanillin | 1 × 10−8 | Si-ISFET | This work |
| Protein molecules | |||
| Thrombin | 2.5 × 10−8 | Si-ISFET | [42] |
| Thrombin | 7 × 10−7 | Si-ISFET | [43] |
| Thrombin | 5 × 10−8 | Polypyrrole-FET | [44] |
| Thrombin | 2 × 10−11 | Carbon-FET | [45] |
| Vascular endotherial growth factor | 1.04 × 10−9–1.04 × 10−10 | Si-ISFET | [46] |
| IgE | Kdiss = 4.7 × 10−8 | Graphene-ISFET | [47] |
| IgE | 2.5 × 10−10 | Carbon-FET | [48] |
| Interferon gamma | 8.3 × 10−11 | Graphene-ISFET | [49] |
| Platelet-derived growth factor | 5 × 10−12 | Carbon-FET | [50] |
| Platelet-derived growth factor | - | Diamond-FET | [51] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrianova, M.; Komarova, N.; Grudtsov, V.; Kuznetsov, E.; Kuznetsov, A. Amplified Detection of the Aptamer–Vanillin Complex with the Use of Bsm DNA Polymerase. Sensors 2018, 18, 49. https://doi.org/10.3390/s18010049
Andrianova M, Komarova N, Grudtsov V, Kuznetsov E, Kuznetsov A. Amplified Detection of the Aptamer–Vanillin Complex with the Use of Bsm DNA Polymerase. Sensors. 2018; 18(1):49. https://doi.org/10.3390/s18010049
Chicago/Turabian StyleAndrianova, Mariia, Natalia Komarova, Vitaliy Grudtsov, Evgeniy Kuznetsov, and Alexander Kuznetsov. 2018. "Amplified Detection of the Aptamer–Vanillin Complex with the Use of Bsm DNA Polymerase" Sensors 18, no. 1: 49. https://doi.org/10.3390/s18010049
APA StyleAndrianova, M., Komarova, N., Grudtsov, V., Kuznetsov, E., & Kuznetsov, A. (2018). Amplified Detection of the Aptamer–Vanillin Complex with the Use of Bsm DNA Polymerase. Sensors, 18(1), 49. https://doi.org/10.3390/s18010049