Significance Testing and Multivariate Analysis of Datasets from Surface Plasmon Resonance and Surface Acoustic Wave Biosensors: Prediction and Assay Validation for Surface Binding of Large Analytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Chemicals
2.1.2. Biomolecules for Acetylcholinesterase/AFB1-Bioconjugates Binding Assays
2.1.3. Biomolecules for Anti-AFB1 Antibody/AFB1-Bioconjugates Binding Assays
2.1.4. Assay Buffers
2.2. Apparatus
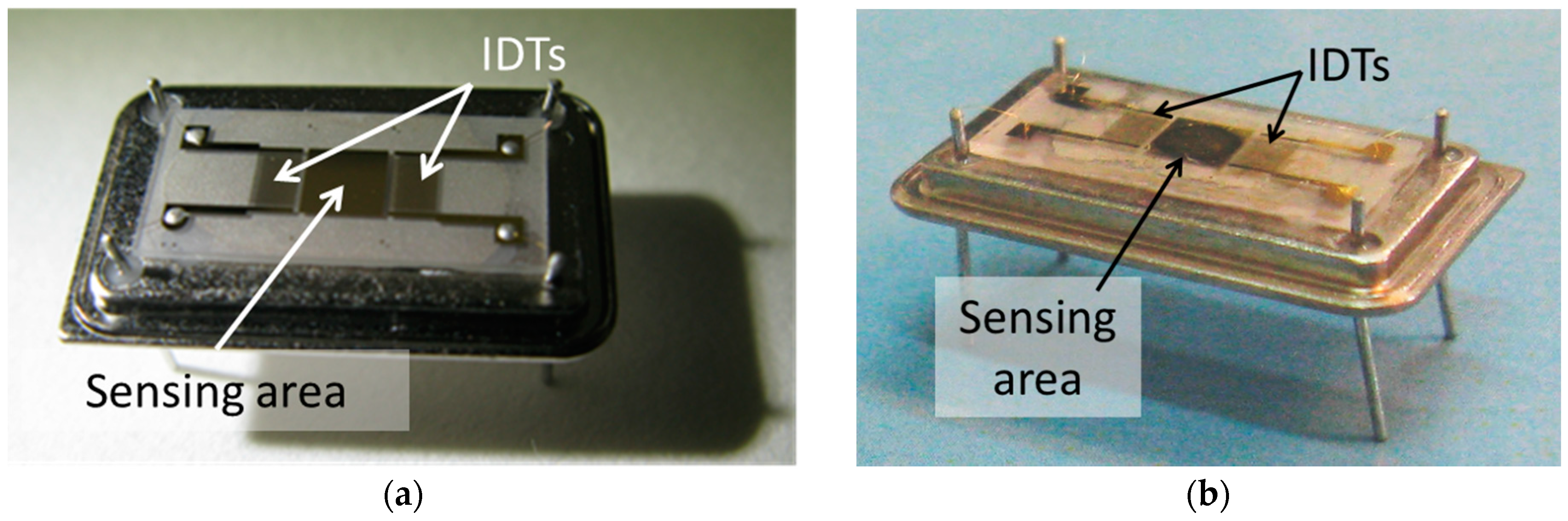
2.2.1. Configurations of the Custom-Made LW-SAW Cells
Piezoelectric Substrates for the LW-SAW Cells
Waveguide Mode and Deposition of the Sensitive Layers
Design of the LW-SAW Sensor Testing Cells
2.3. Gold Surface Coating with a Thiolic Self-Assembled Monolayer
2.4. Immobilization of Biomolecules onto SAM Coated Surfaces
2.4.1. Immobilization of AChE for SPR Binding Assays
- Activation of the carboxylic groups of immobilized 11-MUA via EDC/NHS estersThis was achieved by successively injecting 50 µL of a 1:1 mixture of 0.4 M EDC/0.1 M, three times for 10 min;
- Optimization of the coupling buffer and of the concentration for AChE immobilization onto SAM.AChE solutions were prepared in a 10 mM acetate buffer (pH 4.5). The pH value was chosen below 1 unit of the isoelectric point of AChE (pI = 5.5) to avoid repulsive interactions between the negatively charged carboxyl groups of 11-MUA and the negatively charged enzyme; AChE was immobilized by injecting 50 µL of solution of 1 µM AChE (protein concentration) for 20 min;
- Blocking of the remaining active sitesThis step was achieved by injection of 50 µL of 1 M ethanolamine (pH 8.5) for 20 min.
2.4.2. Immobilization of Anti-AFB1 Antibody for SPR and SAW Assays
2.5. Monitoring AChE/AFB1-HRP and AChE/AFB1-BSA Binding through SPR Measurements
2.6. Monitoring Anti-AFB1 Antibody/AFB1-BSA Interaction through SPR and LWSAW Measurements
2.6.1. Binding Protocol for SPR Cells Operating in Batch Configuration
2.6.2. Binding Protocol for LW-SAW Cells Operating in Batch Configuration
3. Results and Discussion
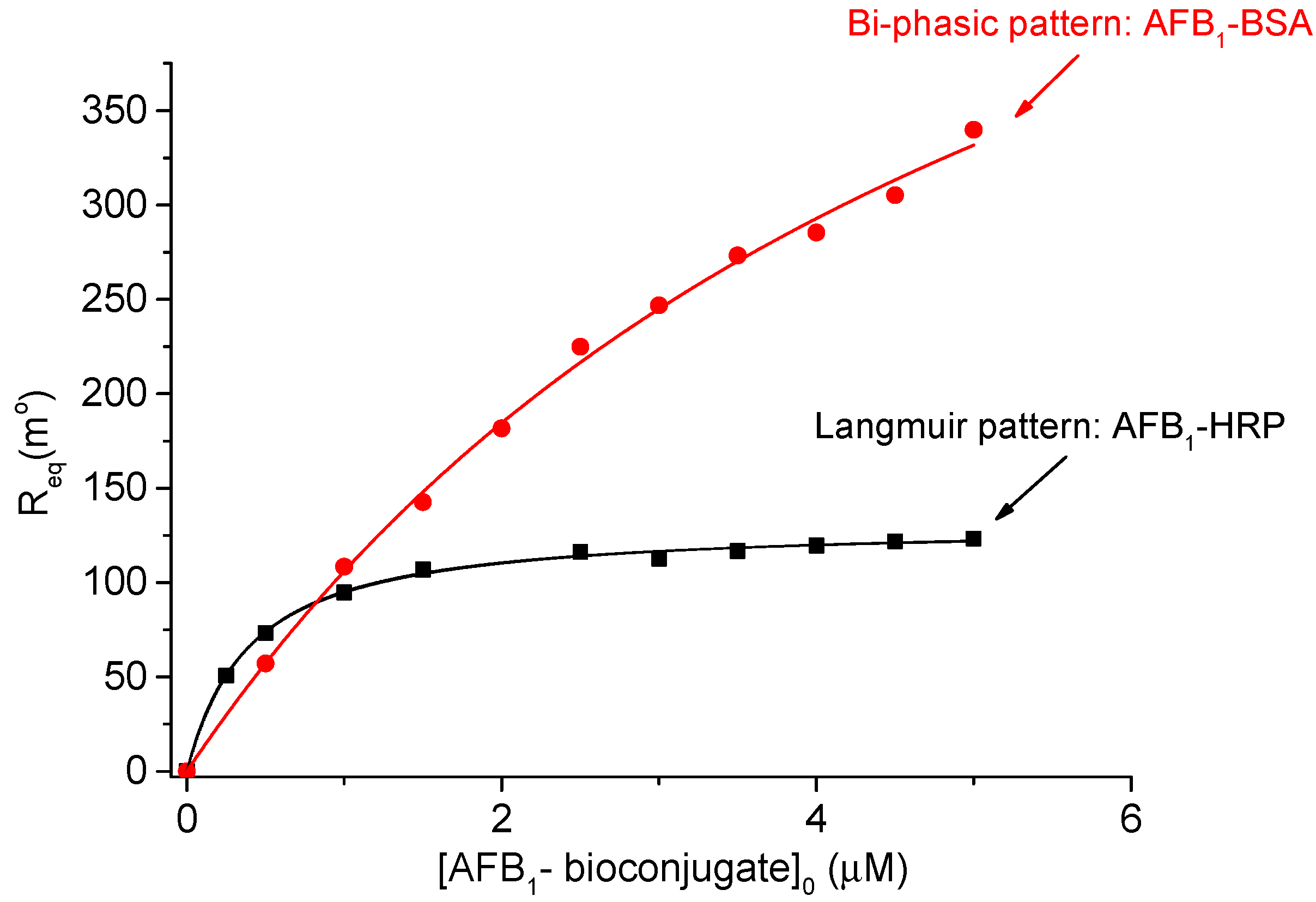
3.1. Affinity and Kinetic Analysis of AChE/AFB1-HRP and AChE/AFB1-BSA Interactions
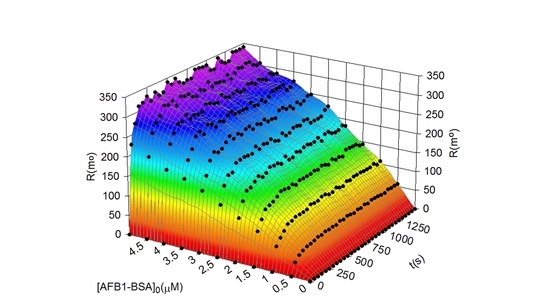
Prediction and Simulation of the SPR Response Using the Bi-Phasic Model
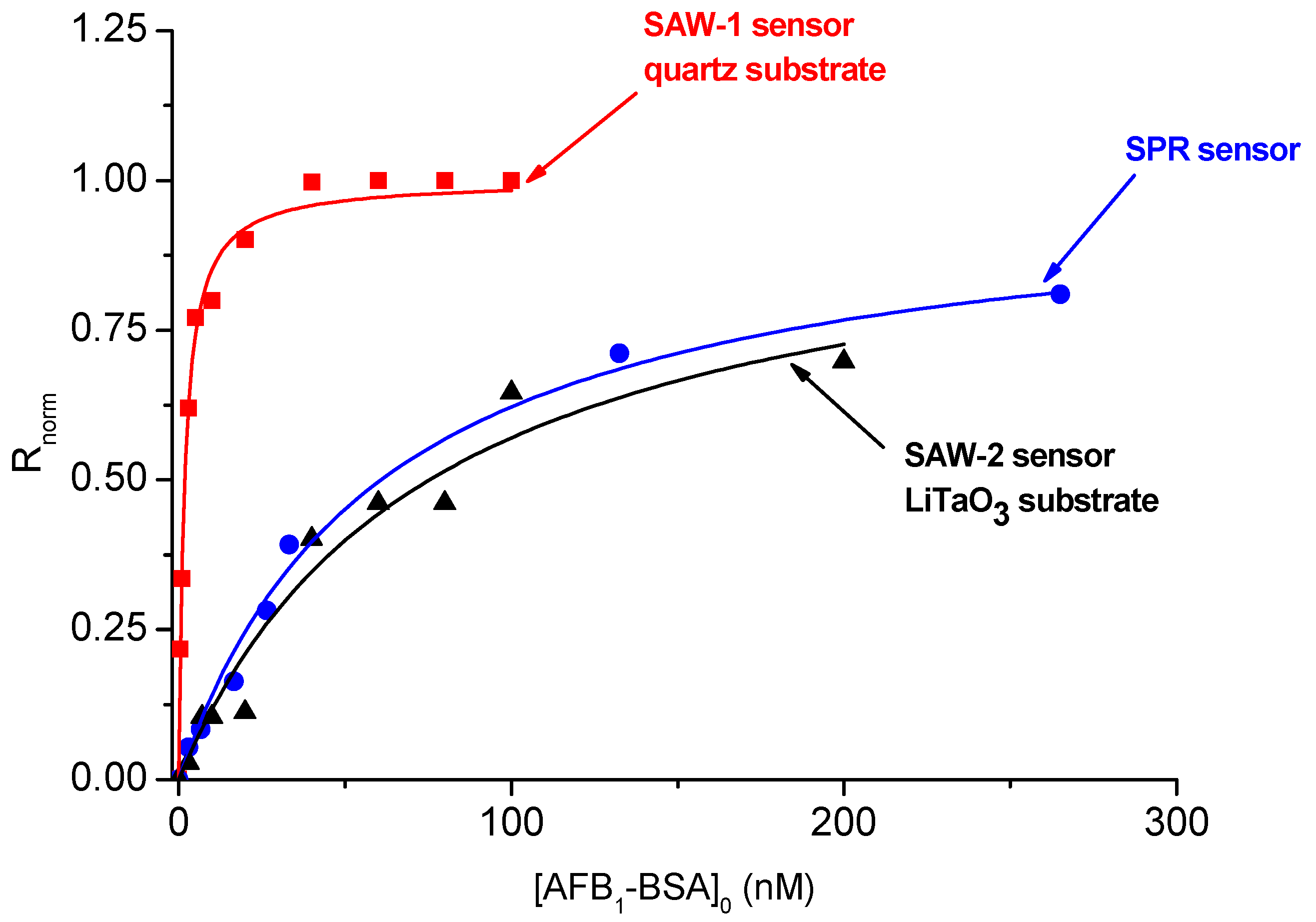
3.2. Testing and Calibrating the LW-SAW Sensors Using Anti-AFB1 Antibody/AFB1-BSA Affinity Pair
3.3. Analysis and Validation of Data Using the F- and t-Tests for Unpaired Data
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Puiu, M.; Idili, A.; Moscone, D.; Ricci, F.; Bala, C. A modular electrochemical peptide-based sensor for antibody detection. Chem. Commun. 2014, 50, 8962–8965. [Google Scholar] [CrossRef] [PubMed]
- Puiu, M.; Bala, C. SPR and SPR Imaging: Recent Trends in Developing Nanodevices for Detection and Real-Time Monitoring of Biomolecular Events. Sensors 2016, 16, 870. [Google Scholar] [CrossRef] [PubMed]
- Puiu, M.; Gurban, A.-M.; Rotariu, L.; Brajnicov, S.; Viespe, C.; Bala, C. Enhanced Sensitive Love Wave Surface Acoustic Wave Sensor Designed for Immunoassay Formats. Sensors 2015, 15, 10511–10525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, H.; Park, J.; Kang, S.; Kim, M. Surface Plasmon Resonance: A Versatile Technique for Biosensor Applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helmerhorst, E.; Chandler, D.J.; Nussio, M.; Mamotte, C.D. Real-time and Label-free Bio-sensing of Molecular Interactions by Surface Plasmon Resonance: A Laboratory Medicine Perspective. Clin. Biochem. Rev. 2012, 33, 161–173. [Google Scholar] [PubMed]
- Gronewold, T.M.A. Surface acoustic wave sensors in the bioanalytical field: Recent trends and challenges. Anal. Chim. Acta 2007, 603, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Toma, K.; Miki, D.; Kishikawa, C.; Yoshimura, N.; Miyajima, K.; Arakawa, T.; Yatsuda, H.; Mitsubayashi, K. Repetitive Immunoassay with a Surface Acoustic Wave Device and a Highly Stable Protein Monolayer for On-Site Monitoring of Airborne Dust Mite Allergens. Anal. Chem. 2015, 87, 10470–10474. [Google Scholar] [CrossRef] [PubMed]
- Mujahid, A.; Dickert, F. Surface Acoustic Wave (SAW) for Chemical Sensing Applications of Recognition Layers. Sensors 2017, 17, 2716. [Google Scholar] [CrossRef] [PubMed]
- Najla Fourati, C.Z. Immunosensing with Surface Acoustic Wave Sensors: Toward Highly Sensitive and Selective Improved Piezoelectric Biosensors. In New Sensors and Processing Chain; Jean-Hugh Thomas, N.Y., Ed.; Wiley: New York, NY, USA, 2014; pp. 35–45. [Google Scholar]
- Puiu, M.; Istrate, O.; Rotariu, L.; Bala, C. Kinetic approach of aflatoxin B1–acetylcholinesterase interaction: A tool for developing surface plasmon resonance biosensors. Anal. Biochem. 2012, 421, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Schasfoort, R.B.M. Handbook of Surface Plasmon Resonance, 2nd ed.; Royal Society of Chemistry: Cambridge, UK, 2017; pp. 133–147. [Google Scholar]
- Blondeau-Patissier, V.; Boireau, W.; Cavallier, B.; Lengaigne, G.; Daniau, W.; Martin, G.; Ballandras, S. Integrated Love Wave Device Dedicated to Biomolecular Interactions Measurements in Aqueous Media. Sensors 2007, 7, 1992–2003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindner, M.; Schmid, M. Thickness Measurement Methods for Physical Vapor Deposited Aluminum Coatings in Packaging Applications: A Review. Coatings 2017, 7, 9. [Google Scholar] [CrossRef]
- Fischer, M.J.E. Amine Coupling Through EDC/NHS: A Practical Approach. In Surface Plasmon Resonance: Methods and Protocols; Mol, N.J., Fischer, M.J.E., Eds.; Humana Press: Totowa, NJ, USA, 2010; pp. 55–73. [Google Scholar]
- Boyd, A.E.; Dunlop, C.S.; Wong, L.; Radić, Z.; Taylor, P.; Johnson, D.A. Nanosecond Dynamics of Acetylcholinesterase Near the Active Center Gorge. J. Biol. Chem. 2004, 279, 26612–26618. [Google Scholar] [CrossRef] [PubMed]
- Rich, R.L.; Myszka, D.G. Advances in surface plasmon resonance biosensor analysis. Curr. Opin. Biotechnol. 2000, 11, 54–61. [Google Scholar] [CrossRef]
- Myszka, D.G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr. Opin. Biotechnol. 1997, 8, 50–57. [Google Scholar] [CrossRef]
- Zhao, H.; Gorshkova, I.I.; Fu, G.L.; Schuck, P. A comparison of binding surfaces for SPR biosensing using an antibody–antigen system and affinity distribution analysis. Methods 2013, 59, 328–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusling, J.F.; Kumosinski, T.F. Nonlinear Computer Modeling of Chemical and Biochemical Data; Elsevier Science: Amsterdam, The Netherlands, 1996; pp. 10–25. [Google Scholar]
- Harvey, D. Modern Analytical Chemistry; McGraw-Hill: New York, NY, USA, 2000; pp. 82–96. [Google Scholar]



| k1on (M−1 s−1) | k1off∙103 (s−1) | k2on (M−1 s−1) | k2off ∙103 (s−1) | R1max (m°) | R2max (m°) | Rmax estim (m°) | Rmax exp (m°) | r2 |
|---|---|---|---|---|---|---|---|---|
| 2998 ± 248 | 9.65 ± 0.61 | 1785 ± 121 | 6.54 ± 0.51 | 380 ± 22 | 145 ± 10 | 525 ± 32 | 530 ± 29 | 0.9972 |
| [AFB1-BSA] (M) Injection | Time (s) | Rexp (m°) | [AFB1-BSA] (M) Predicted |
|---|---|---|---|
| 3.325 × 10−7 | 250 | 102 ± 9.1 | (3.39 ± 0.24) × 10−7 |
| 3.325 × 10−7 | 300 | 120 ± 9.5 | (3.401 ± 0.26) × 10−7 |
| Sensor/Interrogation Mode | Parameter | r2 | Linear Range (nM) | Sensitivity (n.r.u/nM) | LOD (nM) |
|---|---|---|---|---|---|
| SAW-1/Phase | 0.5738 ± 0.048 | 0.9897 | 0.3–4 | 0.1491 ± 0.0101 | 0.2 |
| SAW-2/Phase | 0.0133 ± 0.0012 | 0.9627 | 5–30 | 0.0093 ± 0.0011 | 8 |
| SPR /Angle | 0.01645 ± 0.00101 | 0.9917 | 5–30 | 0.01033 ± 0.00016 | 10 |
| Sensor | Average Rnorm | Average Concentration of AFB1-BSA (nM) | Variance (s2) (nM) | No. of Replicates (n) |
|---|---|---|---|---|
| SAW-2—phase interrogation | 0.118 | 10.14 | 1.211 | 5 |
| SPR—angle interrogation | 0.148 | 9.98 | 0.972 | 6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puiu, M.; Zamfir, L.-G.; Buiculescu, V.; Baracu, A.; Mitrea, C.; Bala, C. Significance Testing and Multivariate Analysis of Datasets from Surface Plasmon Resonance and Surface Acoustic Wave Biosensors: Prediction and Assay Validation for Surface Binding of Large Analytes. Sensors 2018, 18, 3541. https://doi.org/10.3390/s18103541
Puiu M, Zamfir L-G, Buiculescu V, Baracu A, Mitrea C, Bala C. Significance Testing and Multivariate Analysis of Datasets from Surface Plasmon Resonance and Surface Acoustic Wave Biosensors: Prediction and Assay Validation for Surface Binding of Large Analytes. Sensors. 2018; 18(10):3541. https://doi.org/10.3390/s18103541
Chicago/Turabian StylePuiu, Mihaela, Lucian-Gabriel Zamfir, Valentin Buiculescu, Angela Baracu, Cristina Mitrea, and Camelia Bala. 2018. "Significance Testing and Multivariate Analysis of Datasets from Surface Plasmon Resonance and Surface Acoustic Wave Biosensors: Prediction and Assay Validation for Surface Binding of Large Analytes" Sensors 18, no. 10: 3541. https://doi.org/10.3390/s18103541