Detection of Metallothionein in Javanese Medaka (Oryzias javanicus), Using a scFv-Immobilized Protein Chip
Abstract
1. Introduction
2. Materials and Methods
2.1. General Procedures
2.2. Purification of Anti-OjaMT mAbs
2.3. Expression and Purification of Recombinant OjaMT
2.4. Immunoblot Analysis
2.5. Isolation of the Variable Heavy and Light Chain (VH and VL)
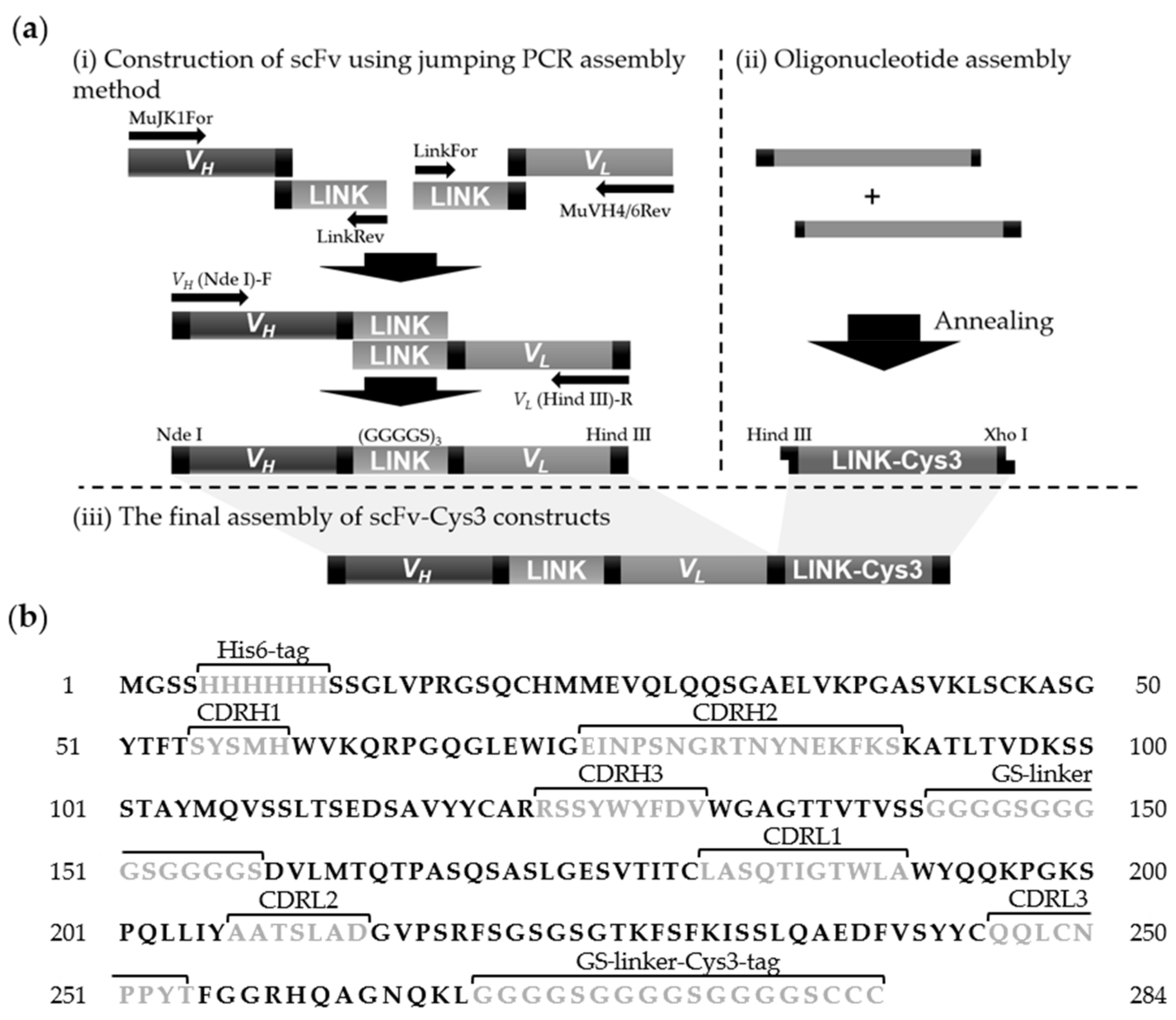
2.6. Construction of Cys3-Tagged scFv (scFv-Cys3)
2.7. Expression and Purification of Recombinant scFv
2.8. Experimental Fish
2.9. CdCl2 Exposure and Sampling (Heavy Metal Exposure and Sampling)
2.10. SPR Analysis
3. Results and Discussion
3.1. Preparation of OjaMT Detection Probes
3.2. Preparation of mAb Sensor Chip and In Situ Real-Time Detection of OjaMT
3.3. Preparation of scFv Sensor Chip and In Situ Real-Time Detection of OjaMT
3.4. Detection of OjaMT in Liver Samples of Heavy Metal Contaminated Oryzias javanicus Using scFv Sensor Chips
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Naser, H.A. Assessment and management of heavy metal pollution in the marine environment of the Arabian Gulf: A review. Mar. Pollut. Bull. 2013, 72, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yang, S. Heavy metal enrichments in the Changjiang (Yangtze River) catchment and on the inner shelf of the East China Sea over the last 150 years. Sci. Total Environ. 2016, 543, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Sin, S.N.; Chua, H.; Lo, W.; Ng, L.M. Assessment of heavy metal cations in sediments of Shing Mun River, Hong Kong. Environ. Int. 2001, 26, 297–301. [Google Scholar] [CrossRef]
- Farkas, A.; Erratico, C.; Viganò, L. Assessment of the environmental significance of heavy metal pollution in surficial sediments of the River Po. Chemosphere 2007, 68, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.W. The Effects of Heavy Metals (other than Mercury) on Marine and Estuarine Organisms. Proc. R. Soc. Lond. B Biol. Sci. 1971, 177, 389–410. [Google Scholar] [CrossRef] [PubMed]
- Reichelt-Brushett, A.; Hudspith, M. The effects of metals of emerging concern on the fertilization success of gametes of the tropical scleractinian coral Platygyra daedalea. Chemosphere 2016, 150, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Barros, M.P.; Campos, S.C.G.; Pinto, E.; Rajamani, S.; Sayre, R.T. Biochemical biomarkers in algae and marine pollution: A review. Ecotoxicol. Environ. Saf. 2008, 71, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.K.S.; Gray, J.S. The use of biomarkers in environmental monitoring programmes. Mar. Pollut. Bull. 2003, 46, 182–186. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, J.; Fu, J.; Shi, J.; Jiang, G. Biomonitoring: An appealing tool for assessment of metal pollution in the aquatic ecosystem. Anal. Chim. Acta 2008, 606, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Aldo, V.; Burlando, B.; Dondero, F.; Marro, A.; Fabbri, R. Metallothionein as a tool in biomonitoring programmes. Biomarkers 1999, 4, 455–466. [Google Scholar] [CrossRef]
- Olafson, R.W.; McCubbin, W.D.; Kay, C.M. Primary- and secondary-structural analysis of a unique prokaryotic metallothionein from Synechococcus sp. cyanobacterium. Biochem. J. 1988, 251, 691. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.; Yum, S.; Jung, H.J.; Shim, W.J.; Lee, C.H.; Lee, T.K. Heavy Metal-Induced Differential Gene Expression of Metallothionein in Javanese Medaka, Oryzias javanicus. Mar. Biotechnol. 2006, 8, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, D.; Colangelo, C.; Wiliams, K.; Gerstein, M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.; Seilhamer, J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 1997, 18, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Gharib, T.G.; Huang, C.C.; Taylor, J.M.; Misek, D.E.; Kardia, S.L.; Giordano, T.J.; Iannettoni, M.D.; Orringer, M.B.; Hanash, S.M.; et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol. Cell. Proteom. 2002, 1, 304–313. [Google Scholar] [CrossRef]
- Lichtinghagen, R.; Musholt, P.B.; Lein, M.; Römer, A.; Rudolph, B.; Glen, K.; Hauptmann, S.; Schnorr, D.; Loening, S.; Jung, K. Different mRNA and protein expression of matrix metalloproteinases 2 and 9 and tissue inhibitor of metalloproteinases 1 in benign and malignant prostate tissue. Eur. Urol. 2002, 42, 398–406. [Google Scholar] [CrossRef]
- Chu, M.M.; Cuo, Z.Q.; Muto, N.; Itoh, N.; Tanaka, K.; Ren, H.W. Development of ELISA for metallothionein-II allows determination of heavy metal pollution of fresh water. Front. Biosci. 2006, 11, 2113–2122. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nagamine, T.; Nakajima, K. Development of a High Sensitivity ELISA for the Assay of Metallothionein. Curr. Pharm. Biotechnol. 2013, 14, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Dudak, F.C.; Boyaci, I.H. Rapid and label-free bacteria detection by surface plasmon resonance (SPR) biosensors. Biotechnol. J. 2009, 4, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Jung, D.; Jeon, Y.; Jeoung, E.; Kwon, Y. Site-specific and effective immobilization of proteins by Npu DnaE split-intein mediated protein trans-splicing reaction. Biochip J. 2013, 7, 288–294. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Lee, N.; Woo, S.; Ryu, J.-C.; Yum, S. Transcriptomic change as evidence for cadmium-induced endocrine disruption in marine fish model of medaka, Oryzias javanicus. Mol. Cell. Toxicol. 2016, 12, 409–420. [Google Scholar] [CrossRef]
- Grodzki, A.C.; Berenstein, E. Antibody purification: Affinity chromatography-protein A and protein G Sepharose. Methods Mol. Biol. 2010, 588, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Orum, H.; Anderson, P.S.; Oster, A.; Johansen, L.K.; Riise, E.; Bjørnvad, M.; Svendsen, I.; Engberg, J. Efficient method for constructing comprehensive murine Fab antibody libraries displayed on phage. Nucleic Acids Res. 1993, 21, 4491–4498. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.; Han, Z.; Karatan, E.; Mrksich, M.; Kay, B.K. Antibody Arrays Prepared by Cutinase-Mediated Immobilization on Self-Assembled Monolayers. Anal. Chem. 2004, 76, 5713–5720. [Google Scholar] [CrossRef] [PubMed]
- Khodadoust, D.; Ahmad, I. Metallothionein-Like Protein Levels in Java Medaka Fish (Oryzias javanicus) Exposed to Different Concentrations of Cadmium. Walailak J. Sci. Technol. 2014, 11, 883–893. [Google Scholar] [CrossRef]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, E.; Jeon, H.; Kang, C.; Woo, S.; Yum, S.; Kwon, Y. Detection of Metallothionein in Javanese Medaka (Oryzias javanicus), Using a scFv-Immobilized Protein Chip. Sensors 2018, 18, 1069. https://doi.org/10.3390/s18041069
Lee E, Jeon H, Kang C, Woo S, Yum S, Kwon Y. Detection of Metallothionein in Javanese Medaka (Oryzias javanicus), Using a scFv-Immobilized Protein Chip. Sensors. 2018; 18(4):1069. https://doi.org/10.3390/s18041069
Chicago/Turabian StyleLee, Euiyeon, Hyunjin Jeon, Chungwon Kang, Seonock Woo, Seungshic Yum, and Youngeun Kwon. 2018. "Detection of Metallothionein in Javanese Medaka (Oryzias javanicus), Using a scFv-Immobilized Protein Chip" Sensors 18, no. 4: 1069. https://doi.org/10.3390/s18041069
APA StyleLee, E., Jeon, H., Kang, C., Woo, S., Yum, S., & Kwon, Y. (2018). Detection of Metallothionein in Javanese Medaka (Oryzias javanicus), Using a scFv-Immobilized Protein Chip. Sensors, 18(4), 1069. https://doi.org/10.3390/s18041069



