Electronic Detection of DNA Hybridization by Coupling Organic Field-Effect Transistor-Based Sensors and Hairpin-Shaped Probes
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
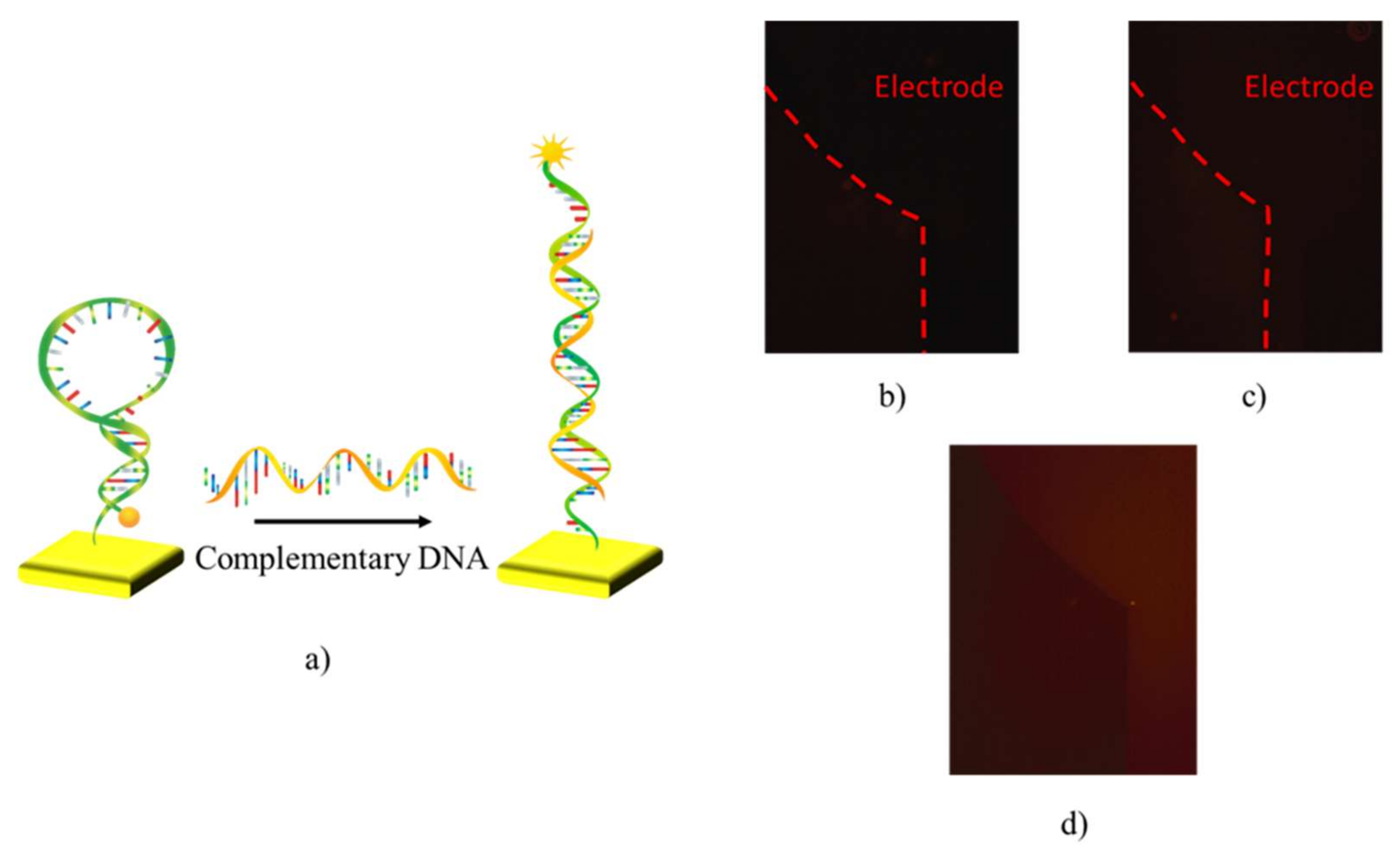
3.1. Functionalization of Sensing Surfaces with Hairpin-Shaped Probes
3.2. Electrical Transduction of DNA Hybridization with Hairpin-Shaped Probes
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
References
- Kaisti, M. Detection principles of biological and chemical FET sensors. Biosens. Bioelectron. 2017, 98, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.; Demelas, M.; Casula, G.; Cosseddu, P.; Barbaro, M.; Bonfiglio, A. Ultralow voltage, OTFT-based sensor for label-free DNA detection. Adv. Mater. 2013, 25, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.; Barbaro, M.; Bonfiglio, A. Tailoring the sensing performances of an OFET-based biosensor. Sens. Actuators B Chem. 2016, 233, 314–319. [Google Scholar] [CrossRef]
- Diculescu, V.C.; Chiorcea-Paquim, A.M.; Oliveira-Brett, A.M. Applications of a DNA-electrochemical biosensor. TrAC Trends Anal. Chem. 2016, 79, 23–36. [Google Scholar] [CrossRef]
- Lucarelli, F.; Tombelli, S.; Minunni, M.; Marrazza, G.; Mascini, M. Electrochemical and piezoelectric DNA biosensors for hybridisation detection. Anal. Chim. Acta 2008, 609, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Ferapontova, E.E. Hybridization Biosensors Relying on Electrical Properties of Nucleic Acids. Electroanalysis 2017, 29, 6–13. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, F.; Ni, J.; Liao, X.; Zhang, X.; Lin, Z. Facile construction of a highly sensitive DNA biosensor by in-situ assembly of electro-active tags on hairpin-structured probe fragment. Sci. Rep. 2016, 6, 22441. [Google Scholar] [CrossRef] [PubMed]
- Farjami, E.; Clima, L.; Gothelf, K.; Ferapontova, E.E. “off-On” electrochemical hairpin-DNA-based genosensor for cancer diagnostics. Anal. Chem. 2011, 83, 1594–1602. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kramer, F.R. Molecular Beacons: Probes that Fluoresce upon Hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, B.; Giannetti, A.; Tombelli, S.; Trono, C.; Baldini, F.; Pellegrino, M.; Sotgiu, G.; Varchi, G. Polymethylmethacrylate nanoparticles as carrier of an oligodeoxynucleotide molecular beacon specific for survivin mRNA in A549 human lung adenocarcinoma epithelial cells. In Proceedings of the 2015 18th AISEM Annual Conference, Trento, Italy, 3–5 February 2015. [Google Scholar]
- Huang, J.; Wu, J.; Li, Z. Biosensing using hairpin DNA probes. Rev. Anal. Chem. 2015, 34, 1–27. [Google Scholar] [CrossRef]
- Tyagi, S.; Kramer, F.R. Molecular beacons in diagnostics. F1000 Med. Rep. 2012, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Ricci, F.; Alexis Vallée-Bélisle, A.; Simon, A.J.; Porchetta, A.; Plaxco, K.W. Using Nature’s “tricks” to Rationally Tune the Binding Properties of Biomolecular Receptors. Acc. Chem. Res. 2016, 49, 1884–1892. [Google Scholar] [CrossRef] [PubMed]
- Farzan, V.M.; Markelov, M.L.; Skoblov, A.Y.; Shipulin, G.A.; Zatsepin, T.S. Specificity of SNP detection with molecular beacons is improved by stem and loop separation with spacers. Analyst 2017, 142, 945–950. [Google Scholar] [CrossRef] [PubMed]
- Stobiecka, M.; Chalupa, A. DNA Strand Replacement Mechanism in Molecular Beacons Encoded for the Detection of Cancer Biomarkers. J. Phys. Chem. B 2016, 120, 4782–4790. [Google Scholar] [CrossRef] [PubMed]
- Demelas, M.; Lai, S.; Casula, G.; Scavetta, E.; Barbaro, M.; Bonfiglio, A. An organic, charge-modulated field effect transistor for DNA detection. Sens. Actuators B Chem. 2012, 171, 198–203. [Google Scholar] [CrossRef]
- Cosseddu, P.; Lai, S.; Barbaro, M.; Bonfiglio, A. Ultra-low voltage, organic thin film transistors fabricated on plastic substrates by a highly reproducible process. Appl. Phys. Lett. 2012, 100, 093305. [Google Scholar] [CrossRef]
- Lai, S.; Cosseddu, P.; Gazzadi, G.C.; Barbaro, M.; Bonfiglio, A. Towards high frequency performances of ultra-low voltage OTFTs: Combining self-alignment and hybrid, nanosized dielectrics. Org. Electron. Phys. Mater. Appl. 2013, 14, 754–761. [Google Scholar] [CrossRef]
- Herne, T.M.; Tarlov, M.J. Characterization of DNA Probes Immobilized on Gold Surfaces. J. Am. Chem. Soc. 1997, 119, 8916–8920. [Google Scholar] [CrossRef]
- Scavetta, E.; Solito, A.G.; Demelas, M.; Cosseddu, P.; Bonfiglio, A. Electrochemical characterization of self assembled monolayers on flexible electrodes. Electrochim. Acta 2012, 65, 159–164. [Google Scholar] [CrossRef]
- Du, H.; Strohsahl, C.M.; Camera, J.; Miller, B.L.; Krauss, T.D. Sensitivity and specificity of metal surface-immobilized “molecular beacon” biosensors. J. Am. Chem. Soc. 2005, 127, 7932–7940. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Disney, M.D.; Miller, B.L.; Krauss, T.D. Hybridization-based unquenching of DNA hairpins on Au surfaces: Prototypical “molecular beacon” biosensors. J. Am. Chem. Soc. 2003, 125, 4012–4013. [Google Scholar] [CrossRef] [PubMed]
- Rant, U.; Arinaga, K.; Fujita, S.; Yokoyama, N.; Abstreiter, G.; Tornow, M. Structural properties of oligonucleotide monolayers on gold surfaces probed by fluorescence investigations. Langmuir 2004, 20, 10086–10092. [Google Scholar] [CrossRef] [PubMed]
- Barbaro, M.; Bonfiglio, A.; Raffo, L.; Alessandrini, A.; Facci, P.; Barák, I. A CMOS, fully integrated sensor for electronic detection of DNA hybridization. IEEE Electron Device Lett. 2006, 27, 595–597. [Google Scholar] [CrossRef]
- Demelas, M.; Lai, S.; Spanu, A.; Martinoia, S.; Cosseddu, P.; Barbaro, M.; Bonfiglio, A. Charge sensing by organic charge-modulated field effect transistors: Application to the detection of bio-related effects. J. Mater. Chem. B 2013, 1, 3811–3819. [Google Scholar] [CrossRef]
- Liu, X.; Tan, W. A fiber-optic evanescent wave DNA biosensor based on novel molecular beacons. Anal. Chem. 1999, 71, 5054–5059. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Tan, W. Molecular-beacon-based array for sensitive DNA analysis. Anal. Biochem. 2004, 331, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Poghossian, A.; Cherstvy, A.; Ingebrandt, S.; Offenhäusser, A.; Schöning, M.J. Possibilities and limitations of label-free detection of DNA hybridization with field-effect-based devices. Sens. Actuators B Chem. 2005, 111, 470–480. [Google Scholar] [CrossRef]
- Lai, S.; Barbaro, M.; Bonfiglio, A. The role of polarization-induced reorientation of DNA strands on organic field-effect transistor-based biosensors sensitivity at high ionic strength. Appl. Phys. Lett. 2015, 107, 103301. [Google Scholar] [CrossRef]
- Rant, U.; Arinaga, K.; Fujita, S.; Yokoyama, N.; Abstreiter, G.; Tornow, M. Electrical manipulation of oligonucleotides grafted to charged surfaces. Org. Biomol. Chem. 2006, 4, 3448–3455. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Napoli, C.; Lai, S.; Giannetti, A.; Tombelli, S.; Baldini, F.; Barbaro, M.; Bonfiglio, A. Electronic Detection of DNA Hybridization by Coupling Organic Field-Effect Transistor-Based Sensors and Hairpin-Shaped Probes. Sensors 2018, 18, 990. https://doi.org/10.3390/s18040990
Napoli C, Lai S, Giannetti A, Tombelli S, Baldini F, Barbaro M, Bonfiglio A. Electronic Detection of DNA Hybridization by Coupling Organic Field-Effect Transistor-Based Sensors and Hairpin-Shaped Probes. Sensors. 2018; 18(4):990. https://doi.org/10.3390/s18040990
Chicago/Turabian StyleNapoli, Corrado, Stefano Lai, Ambra Giannetti, Sara Tombelli, Francesco Baldini, Massimo Barbaro, and Annalisa Bonfiglio. 2018. "Electronic Detection of DNA Hybridization by Coupling Organic Field-Effect Transistor-Based Sensors and Hairpin-Shaped Probes" Sensors 18, no. 4: 990. https://doi.org/10.3390/s18040990
APA StyleNapoli, C., Lai, S., Giannetti, A., Tombelli, S., Baldini, F., Barbaro, M., & Bonfiglio, A. (2018). Electronic Detection of DNA Hybridization by Coupling Organic Field-Effect Transistor-Based Sensors and Hairpin-Shaped Probes. Sensors, 18(4), 990. https://doi.org/10.3390/s18040990







