An Empirical-Mathematical Approach for Calibration and Fitting Cell-Electrode Electrical Models in Bioimpedance Tests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell-Culture Assay
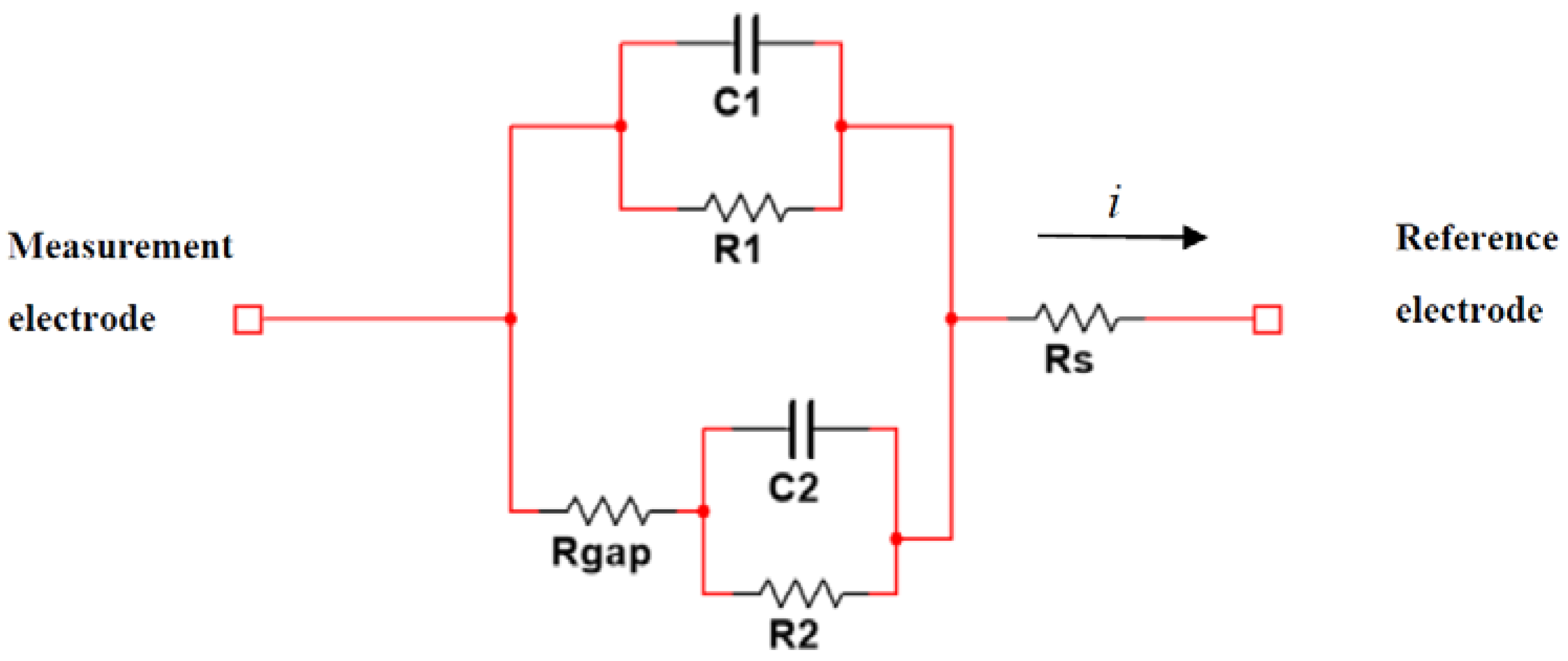
2.2. Cell-Electrode Electrical Model
2.3. Model-Oscillation Relationship
- , at the beginning of the experiment:
- ○
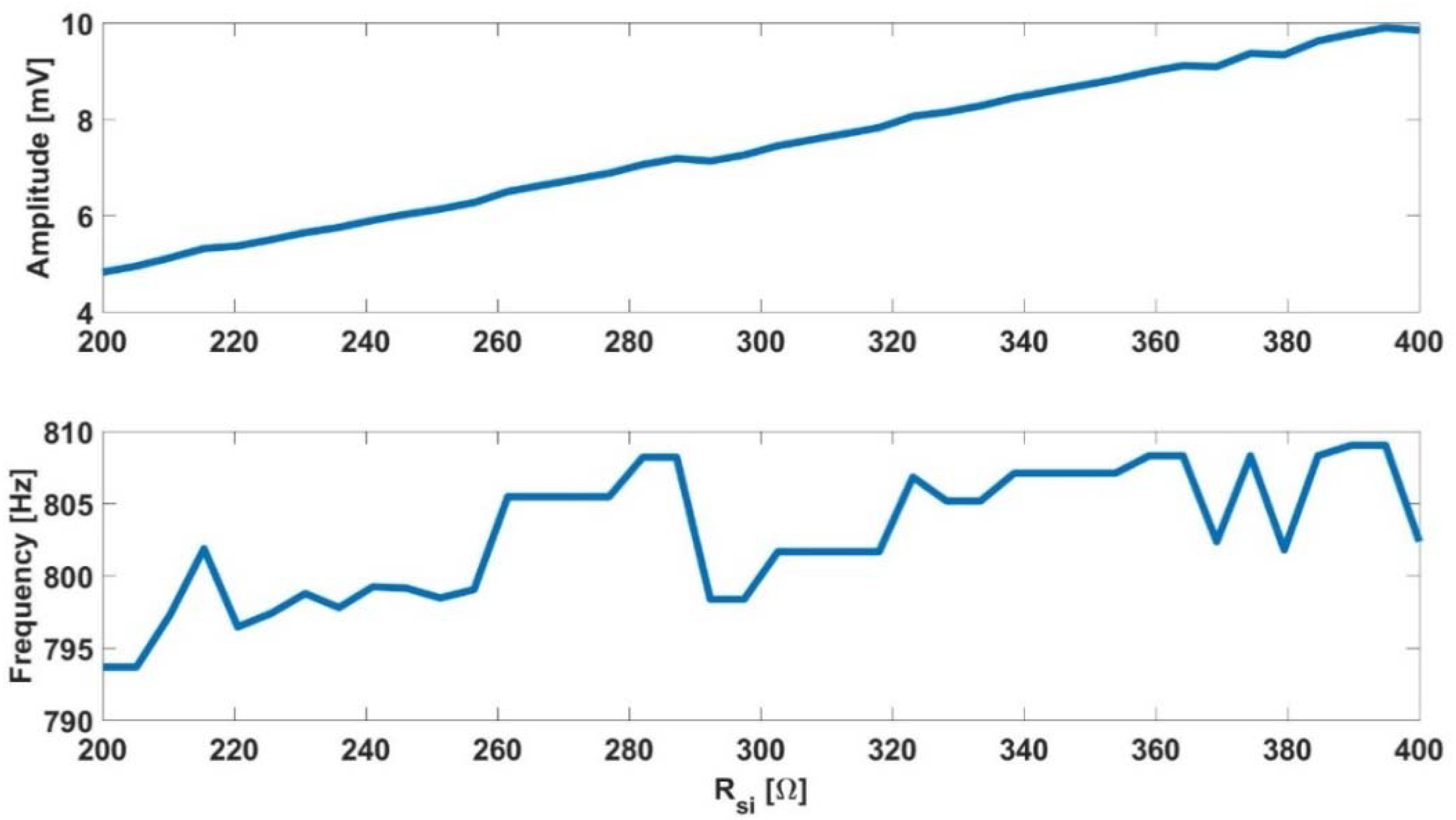
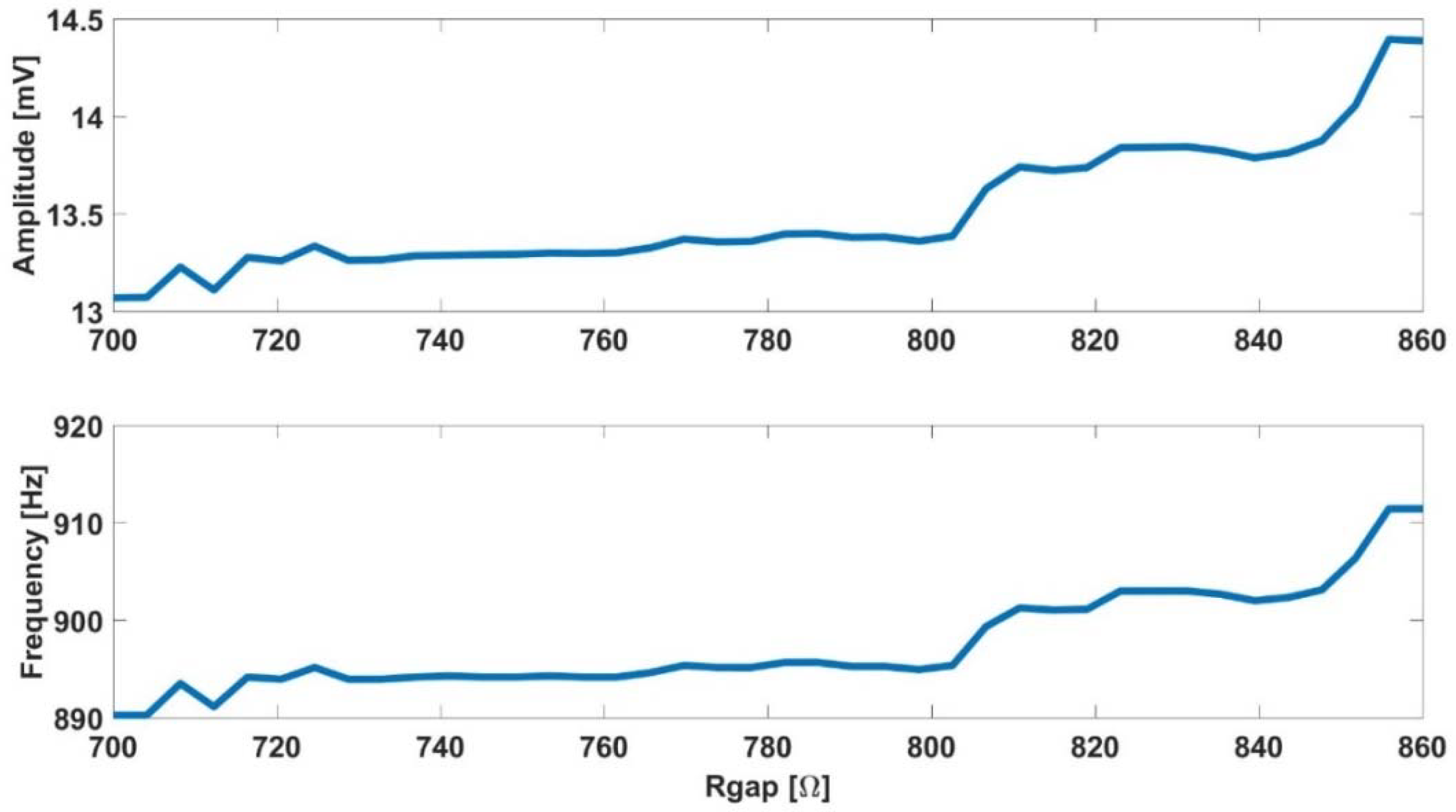
- The initial frequency can be determined only with the position of the pole, .
- ○
- When the pole location is known, the initial amplitude can be determined using the parameter . The resistance effect is significantly higher in initial amplitude than the effect of .
- ○
- , at the end of experiment:
- ○
- The final frequency and amplitude, at the confluent phase, are highly dependent on the parameter.
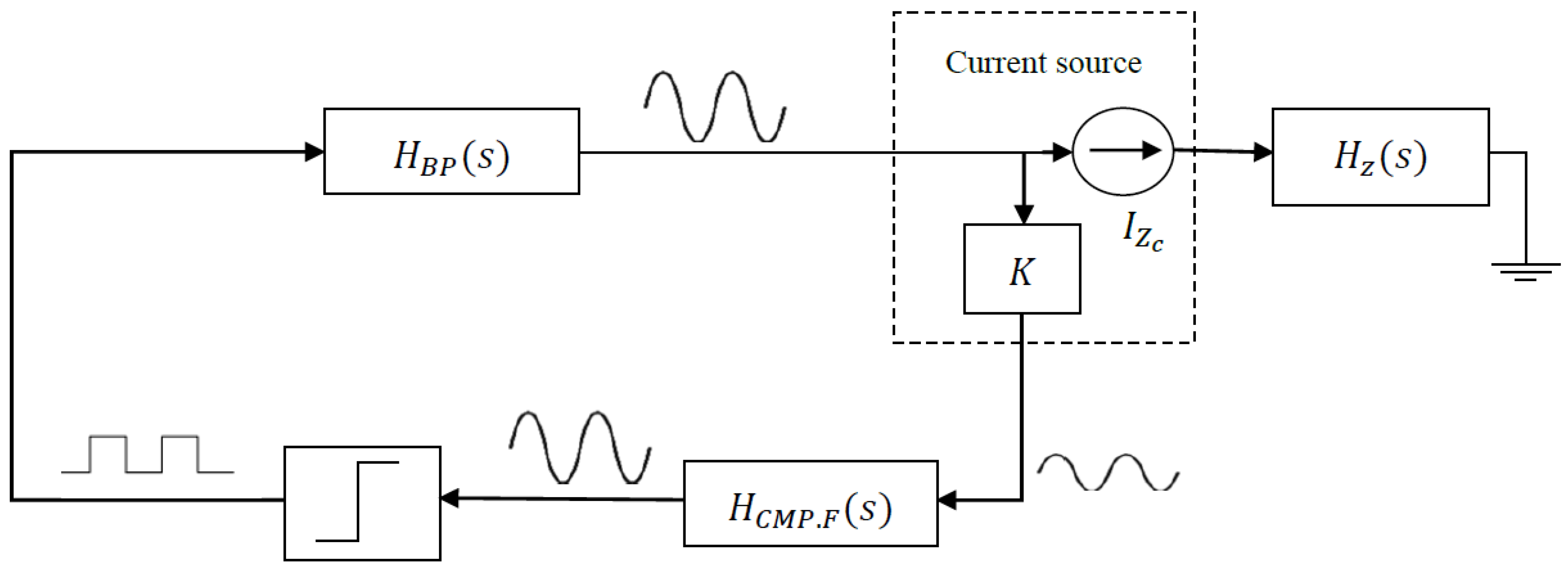
2.4. On-Line Estimation
2.4.1. System Equations
- ○
- The equations of entire system; that is, the equations of the BioZ electric model and the oscillator. From these equations and using the Describing Function method [20], two equations are obtained whose solutions allow us to derive the oscillation amplitude and frequency.
- ○
- The BioZ equations:where is the transfer function of the cell-electrode electrical model and the other parameters are defined by the resistances and the capacitors of the electric model (, , , , and ), as can be seen on Equations (7)–(11).
- ○
- ○
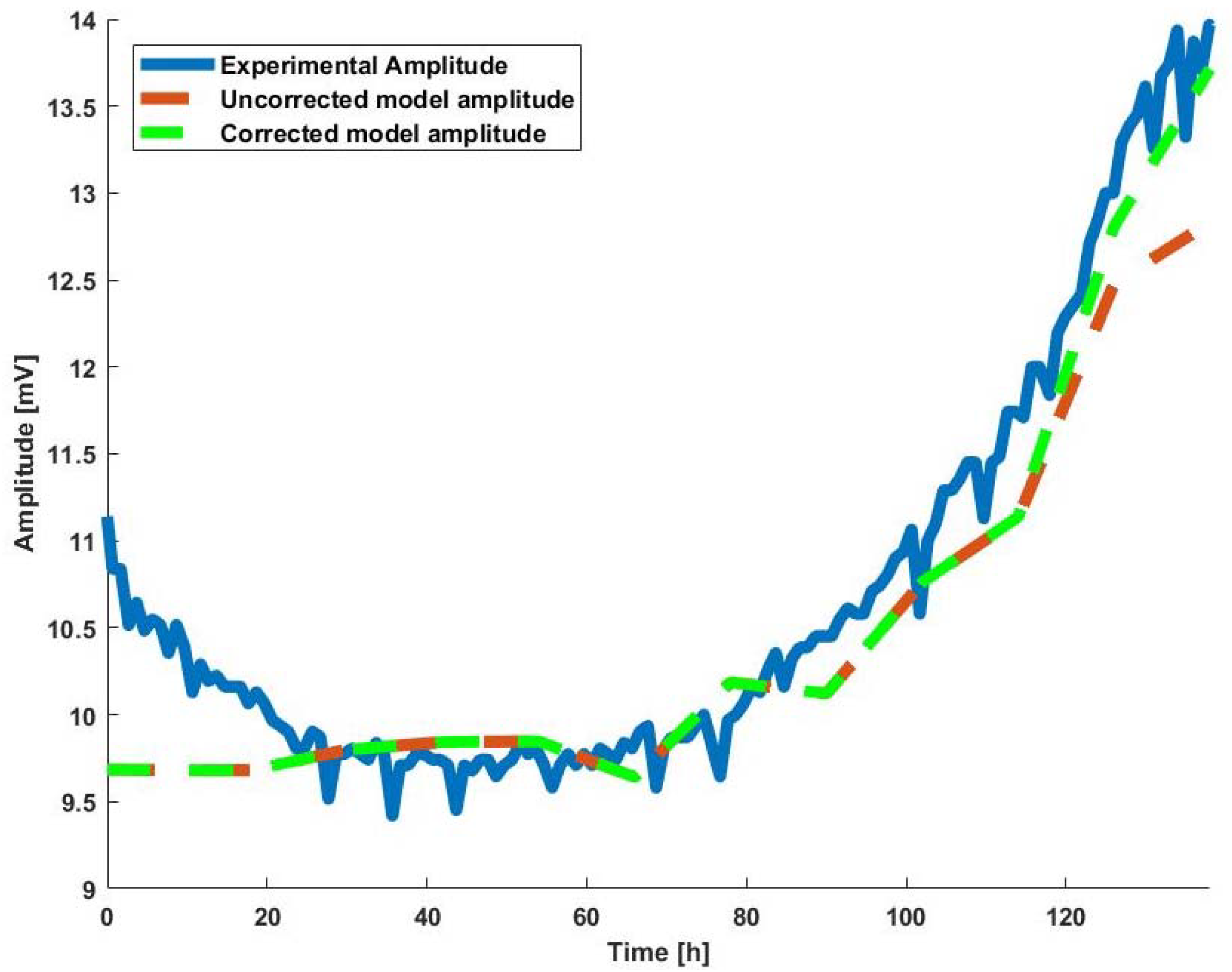
- The parameters of the BioZ electric model, which are obtained from calibration (as will be discussed below), are incorporated to Equation (16), introducing the electrode parameter influence on solutions derived for amplitude and frequency.
- ○
- The relationship between ff and the number of cells, which will depend on the maximum area to be covered by the cells and the cell size for each cell line.
2.4.2. System Calibration
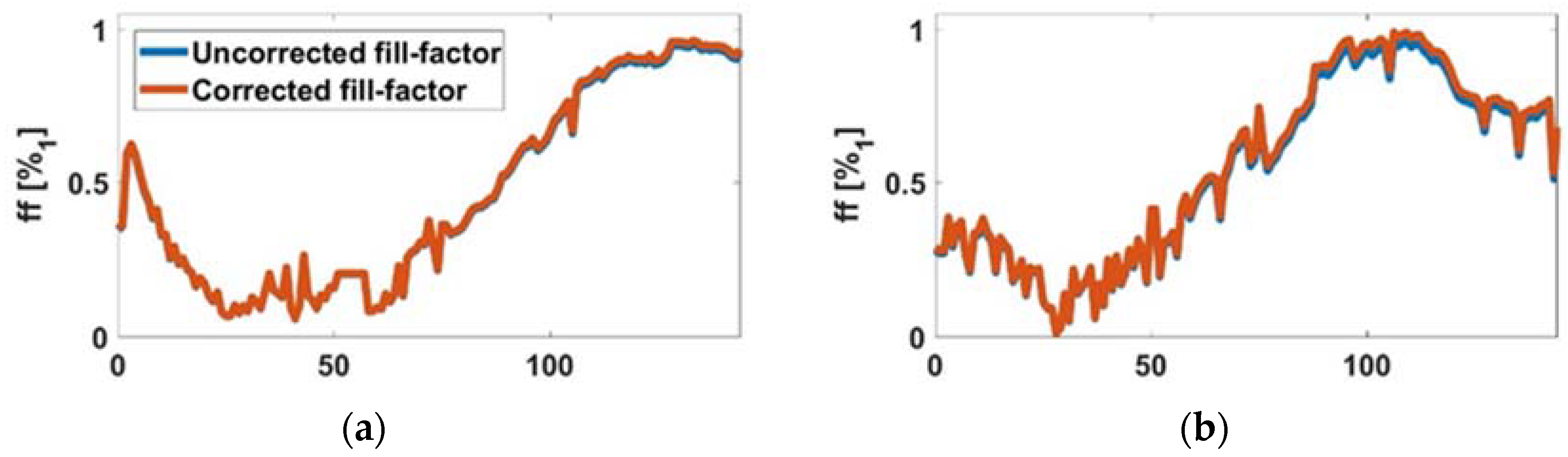
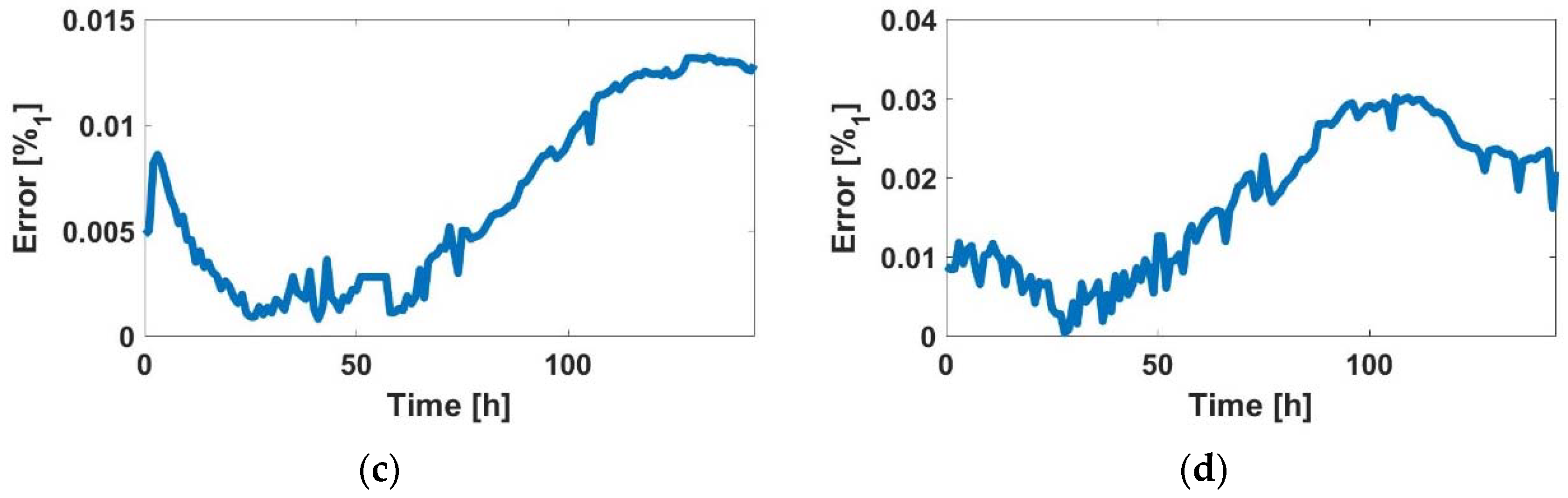
2.4.3. ff-Number of Cells Relationship
3. Results
3.1. Experimental Cell-Line ff Curves
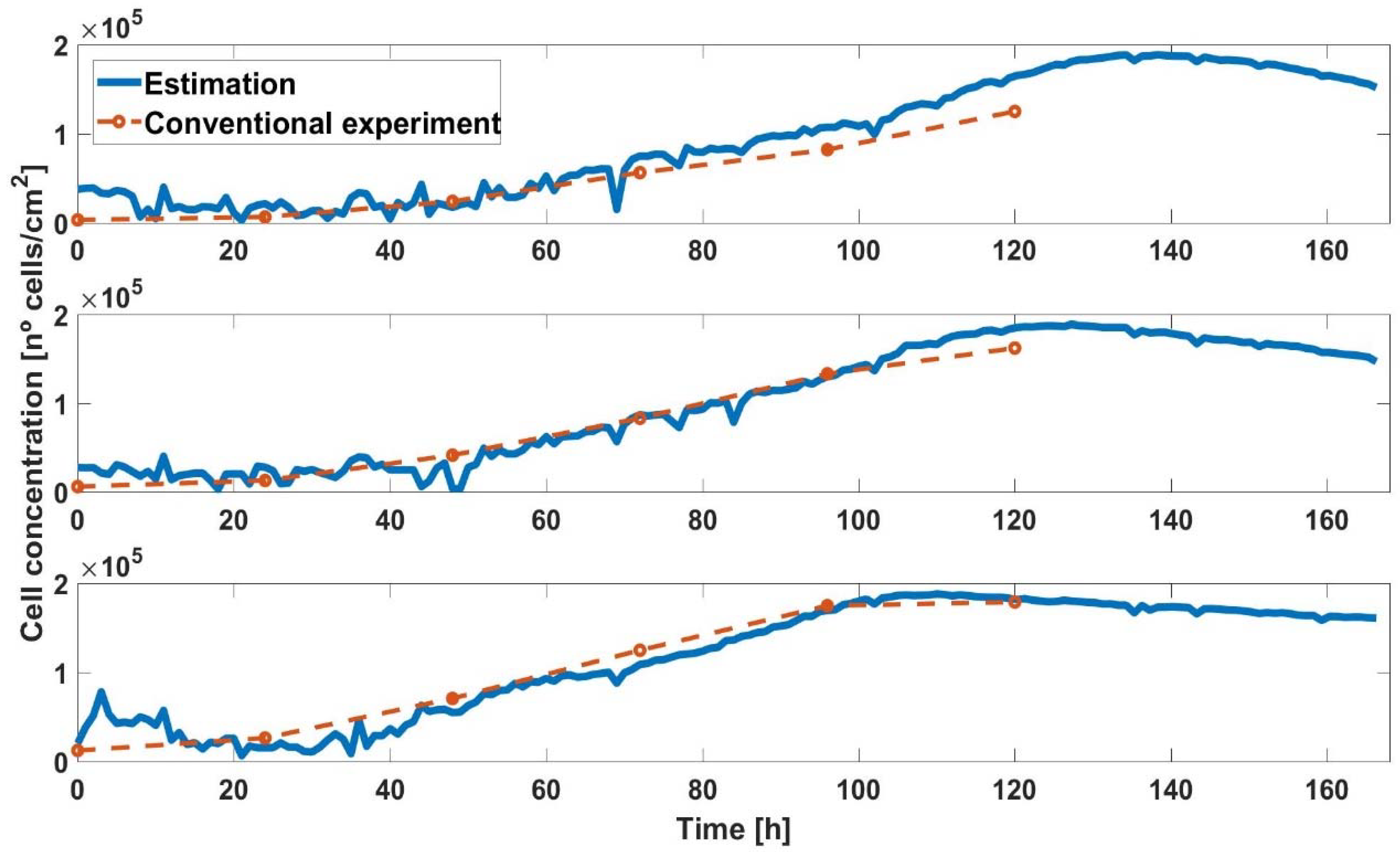
3.2. Experimental Cell-Line Growth Curves
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A




References
- Khalil, S.; Mohktar, M.; Ibrahim, F. The Theory and Fundamentals of Bioimpedance Analysis in Clinical Status Monitoring and Diagnosis of Diseases. Sensors 2014, 14, 10895–10928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.-Y.; Huang, J.-J.; Cheng, K.-S. The design of electrode-array for monitoring the cellular bioimpedance. In Proceedings of the 2009 IEEE Symposium on Industrial Electronics & Applications, Kuala Lumpur, Malaysia, 4–6 October 2009. [Google Scholar]
- Lei, K. Review on Impedance Detection of Cellular Responses in Micro/Nano Environment. Micromachines 2014, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Borkholder, D.A. Cell Based Biosensors Using Microelectrode. Ph.D. Thesis, Stanford University, Stanford, CA, USA, November 1998. [Google Scholar]
- Giaever, I.; Keese, C.R. Use of Electric Fields to Monitor the Dynamical Aspect of Cell Behavior in Tissue Culture. IEEE Trans. Biomed. Eng. 1986, BME-33, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, R.; Mandal, M.; Mitra, A.; Das, S. Monitoring cellular activities of cancer cells using impedance sensing devices. Sens. Actuators B Chem. 2014, 193, 478–483. [Google Scholar] [CrossRef]
- Daza, P.; Olmo, A.; Cañete, D.; Yúfera, A. Monitoring living cell assays with bio-impedance sensors. Sens. Actuators B Chem. 2013, 176, 605–610. [Google Scholar] [CrossRef] [Green Version]
- Abdolahad, M.; Shashaani, H.; Janmaleki, M.; Mohajerzadeh, S. Silicon nanograss based impedance biosensor for label free detection of rare metastatic cells among primary cancerous colon cells, suitable for more accurate cancer staging. Biosens. Bioelectron. 2014, 59, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, C.F.; Ledo, A.; Laranjinha, J.; Gerhardt, G.A.; Barbosa, R.M. Microelectrode array biosensor for high-resolution measurements of extracellular glucose in the brain. Sens. Actuators B Chem. 2016, 237, 298–307. [Google Scholar] [CrossRef]
- Dibao-Dina, A.; Follet, J.; Ibrahim, M.; Vlandas, A.; Senez, V. Electrical impedance sensor for quantitative monitoring of infection processes on HCT-8 cells by the waterborne parasite Cryptosporidium. Biosens. Bioelectron. 2015, 66, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Reitinger, S.; Wissenwasser, J.; Kapferer, W.; Heer, R.; Lepperdinger, G. Electric impedance sensing in cell-substrates for rapid and selective multipotential differentiation capacity monitoring of human mesenchymal stem cells. Biosens. Bioelectron. 2012, 34, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Grimnes, S.; Martinsen, O.G. Bioimpedance and Bioelectricity Basics, 3rd ed.; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Huang, X.; Nguyen, D.; Greve, D.W.; Domach, M.M. Simulation of Microelectrode Impedance Changes Due to Cell Growth. IEEE Sens. J. 2004, 4, 576–583. [Google Scholar] [CrossRef]
- Olmo, A.; Yúfera, A. Computer simulation of microelectrode based bio-impedance measurements with COMSOL. In Proceedings of the Third International Conference on Biomedical Electronics and Devices, Valencia, Spain, 20–23 January 2010; pp. 178–182. [Google Scholar]
- Huertas, G.; Maldonado, A.; Yufera, A.; Rueda, A.; Huertas, J.L. The Bio-Oscillator: A Circuit for Cell-Culture Assays. IEEE Trans. Circuits Syst. II Express Briefs 2015, 62, 164–168. [Google Scholar] [CrossRef] [Green Version]
- Pérez, P.; Maldonado-Jacobi, A.; López, A.J.; Martínez, C.; Olmo, A.; Huertas, G.; Yúfera, A. Remote Sensing of Cell-Culture Assays. In New Insights into Cell Culture Technology; InTech: Rijeka, Croatia, 2017; pp. 135–155. [Google Scholar]
- Serrano, J.A.; Pérez, P.; Maldonado, A.; Martín, M.; Olmo, A.; Daza, P.; Huertas, G.; Yúfera, A. Practical Characterization of Cell-Electrode Electrical Models in Bio-Impedance Assays. In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies, Funchal, Madeira, Portugal, 19–21 January 2018. [Google Scholar]
- Pérez, P.; Huertas, G.; Maldonado-Jacobi, A.; Martín, M.; Serrano, J.A.; Olmo, A.; Daza, P.; Yúfera, A. Sensing Cell-Culture Assays with Low-Cost Circuitry. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Applied Biophysics. Available online: http://www.biophysics.com (accessed on 13 July 2018).
- Huertas, G.; Vázquez García de la Vega, D.; Rueda, A.; Huertas, J.L. Oscillation-Based Test in Mixed-Signal. Circuits; Springer: Dordrecht, The Netherlands, 2006. [Google Scholar]




| Cell Line | |
|---|---|
| AA8 | 531 |
| N2aAPP | 118 |
| N2a | 184 |
| Cell Line | ||
|---|---|---|
| AA8 | 889 | 102 |
| N2aAPP | 572 | 21 |
| N2a | 360 | −19 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano, J.A.; Huertas, G.; Maldonado-Jacobi, A.; Olmo, A.; Pérez, P.; Martín, M.E.; Daza, P.; Yúfera, A. An Empirical-Mathematical Approach for Calibration and Fitting Cell-Electrode Electrical Models in Bioimpedance Tests. Sensors 2018, 18, 2354. https://doi.org/10.3390/s18072354
Serrano JA, Huertas G, Maldonado-Jacobi A, Olmo A, Pérez P, Martín ME, Daza P, Yúfera A. An Empirical-Mathematical Approach for Calibration and Fitting Cell-Electrode Electrical Models in Bioimpedance Tests. Sensors. 2018; 18(7):2354. https://doi.org/10.3390/s18072354
Chicago/Turabian StyleSerrano, Juan A., Gloria Huertas, Andrés Maldonado-Jacobi, Alberto Olmo, Pablo Pérez, María E. Martín, Paula Daza, and Alberto Yúfera. 2018. "An Empirical-Mathematical Approach for Calibration and Fitting Cell-Electrode Electrical Models in Bioimpedance Tests" Sensors 18, no. 7: 2354. https://doi.org/10.3390/s18072354
APA StyleSerrano, J. A., Huertas, G., Maldonado-Jacobi, A., Olmo, A., Pérez, P., Martín, M. E., Daza, P., & Yúfera, A. (2018). An Empirical-Mathematical Approach for Calibration and Fitting Cell-Electrode Electrical Models in Bioimpedance Tests. Sensors, 18(7), 2354. https://doi.org/10.3390/s18072354







