Non-Contact Temperature Control System Applicable to Polymerase Chain Reaction on a Lab-on-a-Disc
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Samples
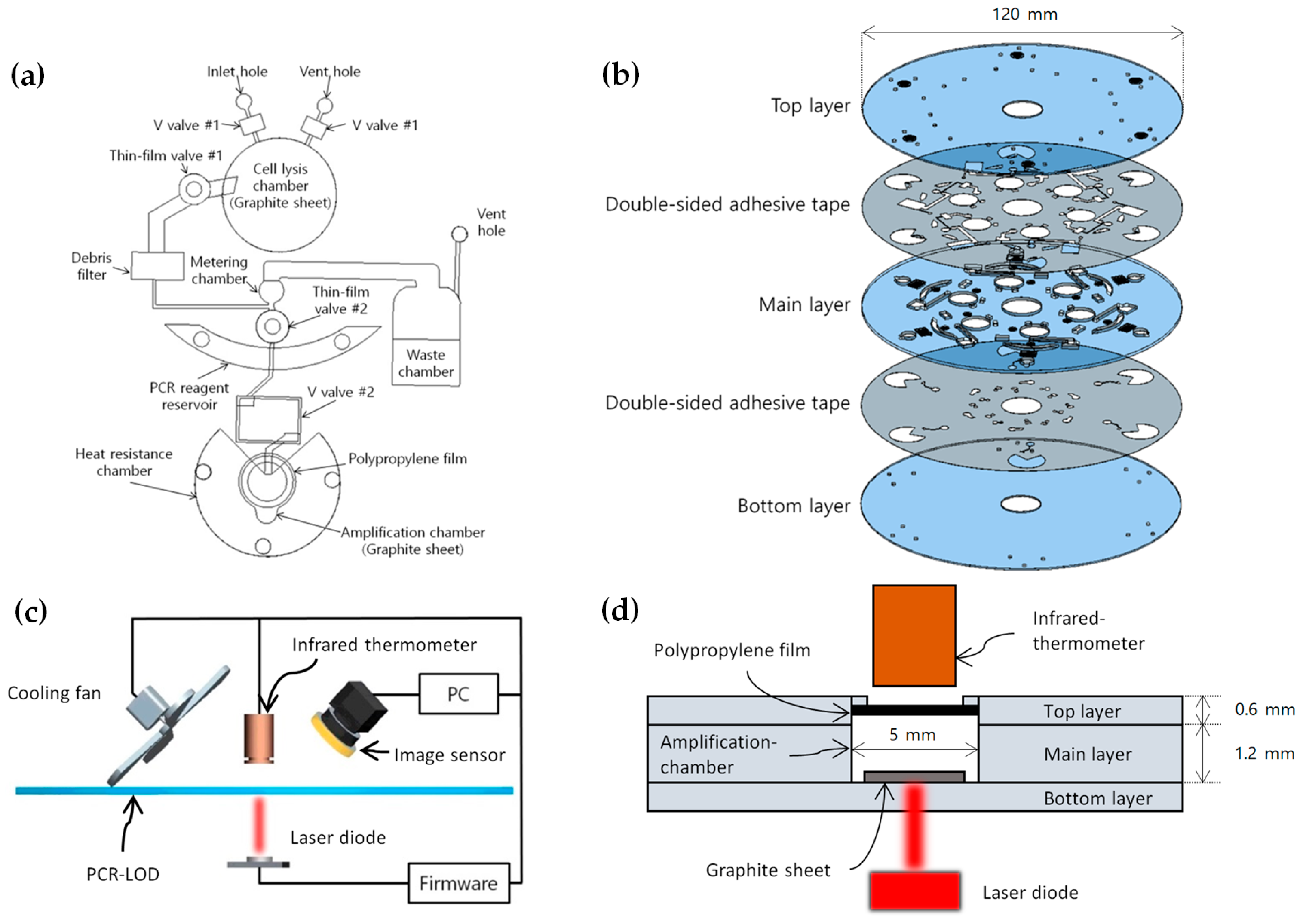
2.2. Design and Fabrication of the Microfluidic Disc
2.3. Temperature Control System
2.4. Microfluidic LOD Operation
2.5. Optical Detection System
3. Results and Discussion
3.1. Sealing of Reagents
3.2. Cell Lysis and DNA Purification
3.3. Temperature Control System for Thermal Cycling
3.4. Optical Detection
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Choi, M.; Yoo, J. Automated centrifugal-microfluidic platform for DNA purification using laser burst valve and coriolis effect. Appl. Biochem. Biotechnol. 2015, 175, 3778–3787. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.; Weber, P.; Lutz, S.; Focke, M.; Zengerle, R.; von Stetten, F. Aliquoting on the centrifugal microfluidic platform based on centrifugo-pneumatic valves. Microfluid. Nanofluid. 2011, 10, 1279–1288. [Google Scholar] [CrossRef]
- Glass, N.R.; Shilton, R.J.; Chan, P.P.; Friend, J.R.; Yeo, L.Y. Miniaturized Lab-on-a-Disc (miniLOAD). Small 2012, 8, 1881–1888. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Sunkara, V.; Kim, T.H.; Hwang, H.; Cho, Y.K. Lab-on-a-disc for fully integrated multiplex immunoassays. Anal. Chem. 2012, 84, 2133–2140. [Google Scholar] [CrossRef] [PubMed]
- Roy, E.; Stewart, G.; Mounier, M.; Malic, L.; Peytavi, R.; Clime, L.; Veres, T. From cellular lysis to microarray detection, an integrated thermoplastic elastomer (TPE) point of care Lab on a Disc. Lab Chip 2015, 15, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ho, H.P.; Chen, Q.; Yang, A.K.L.; Kwok, H.C.; Wu, S.Y.; Zhang, X. A lab-in-a-droplet bioassay strategy for centrifugal microfluidics with density difference pumping, power to disc and bidirectional flow control. Lab Chip 2013, 13, 3698–3706. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M. Infrared Spectroscopy of Pressure-Induced Metallization in Semiconductors. Phys. Status Solidi B 2001, 223, 55–64. [Google Scholar] [CrossRef]
- Combs, L.G.; Warren, J.E.; Huynh, V.; Castaneda, J.; Golden, T.D.; Roby, R.K. The effects of metal ion PCR inhibitors on results obtained with the Quantifiler® Human DNA Quantification Kit. Forensic Sci. Int. Genet. 2015, 19, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Cortés, P.; Spricigo, D.A.; Bardina, C.; Llagostera, M. Remarkable diversity of Salmonella bacteriophages in swine and poultry. FEMS Microbiol. Lett. 2015, 362, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sonnier, R.; Ferry, L.; Gallard, B.; Boudenne, A.; Lavaud, F. Controlled Emissivity Coatings to Delay Ignition of Polyethylene. Materials 2015, 8, 6935–6949. [Google Scholar] [CrossRef] [PubMed]
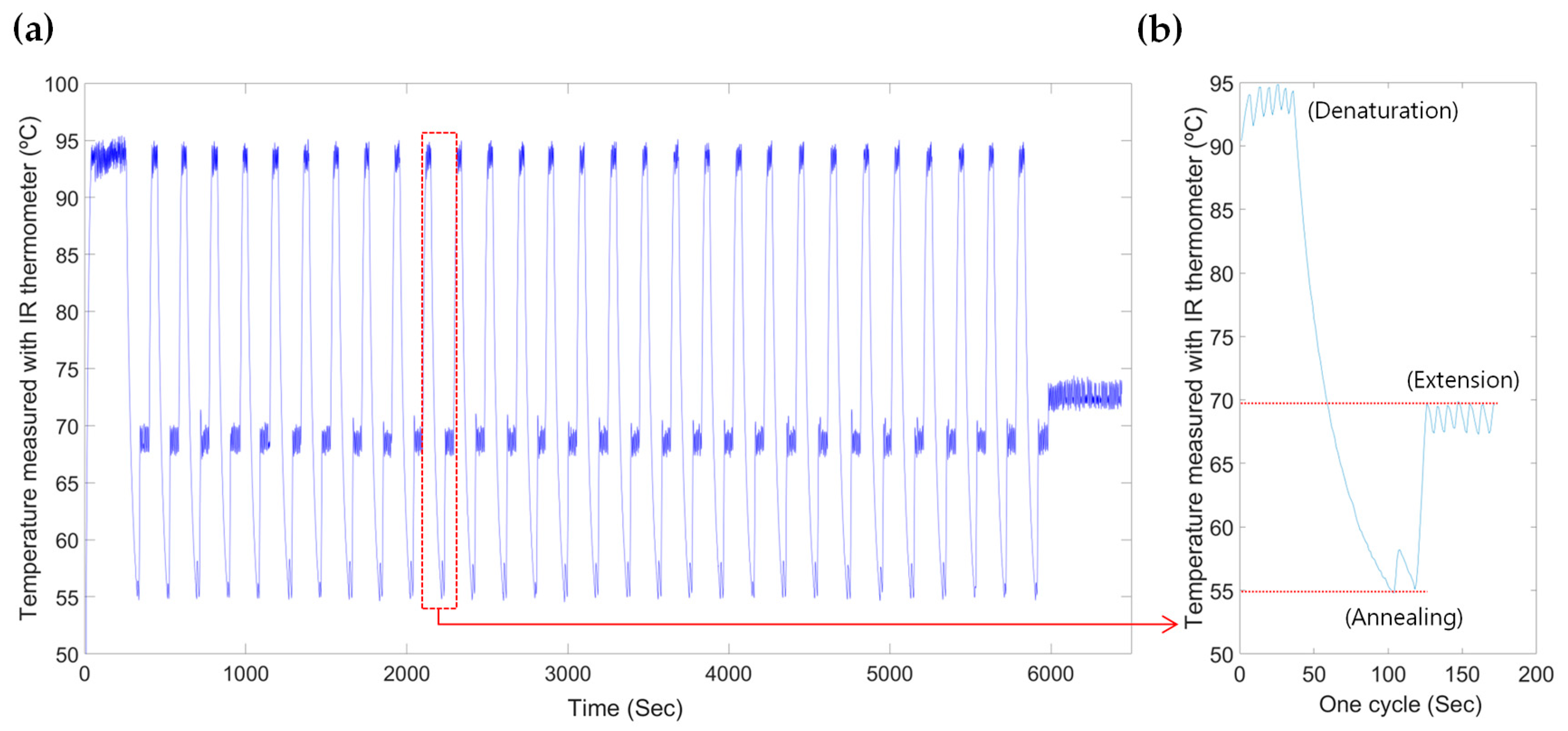




| Step | PCR Phase | Temperature (°C) | Time (s) | Cycle |
|---|---|---|---|---|
| 1 | Initial denaturation | 94 | 120 | 1 |
| 2 | Denaturation | 94 | 20 | 30 |
| 3 | Annealing | 55 | 10 | 30 |
| 4 | Extension | 69 | 30 | 30 |
| 5 | Final extension | 72 | 300 | 1 |
| Step | Procedure | Spin Speed (rpm) | Time (s) |
|---|---|---|---|
| 1 | Load the cultured Salmonella, which was dissolved in the Simplex reagent for cell lysis via the inlet hole | - | - |
| 2 | Close V valve no. 1 with a laser module to seal the lysis chamber | - | 10 |
| 3 | Heat the lysis chamber with a laser module to accelerate the lysis reaction | - | 180 |
| 4 | Spin the disc counterclockwise (CCW) (the reagent is separated into the supernatant containing the purified DNA) | 10,000 | 60 |
| 5 | Open the thin-film valve no. 1 | - | 10 |
| 6 | Spin the disc clockwise (CW) (the supernatant flows through the debris filter and reaches the metering chamber) | 3000 | 10 |
| 7 | Load the PCR premix into the PCR reagent reservoir | - | - |
| 8 | Open the thin-film valve no. 2 | - | 10 |
| 9 | Spin the disc CW (the PCR reagents, now blended with the purified DNA template, flow through the V valve no. 2 and reach the amplification chamber) | 3000 | 10 |
| 10 | Close the V valve no. 2 with a laser module to seal the amplification chamber | - | 10 |
| 11 | Heat the amplification chamber according to the thermal cycles | - | 6400 |
| 12 | Observe the fluorescence of the amplicon excited using a blue LED | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ko, J.; Yoo, J.-C. Non-Contact Temperature Control System Applicable to Polymerase Chain Reaction on a Lab-on-a-Disc. Sensors 2019, 19, 2621. https://doi.org/10.3390/s19112621
Ko J, Yoo J-C. Non-Contact Temperature Control System Applicable to Polymerase Chain Reaction on a Lab-on-a-Disc. Sensors. 2019; 19(11):2621. https://doi.org/10.3390/s19112621
Chicago/Turabian StyleKo, Junguk, and Jae-Chern Yoo. 2019. "Non-Contact Temperature Control System Applicable to Polymerase Chain Reaction on a Lab-on-a-Disc" Sensors 19, no. 11: 2621. https://doi.org/10.3390/s19112621
APA StyleKo, J., & Yoo, J.-C. (2019). Non-Contact Temperature Control System Applicable to Polymerase Chain Reaction on a Lab-on-a-Disc. Sensors, 19(11), 2621. https://doi.org/10.3390/s19112621




