Development of a Simple Assay Method for Adenosine Deaminase via Enzymatic Formation of an Inosine-Tb3+ Complex
Abstract
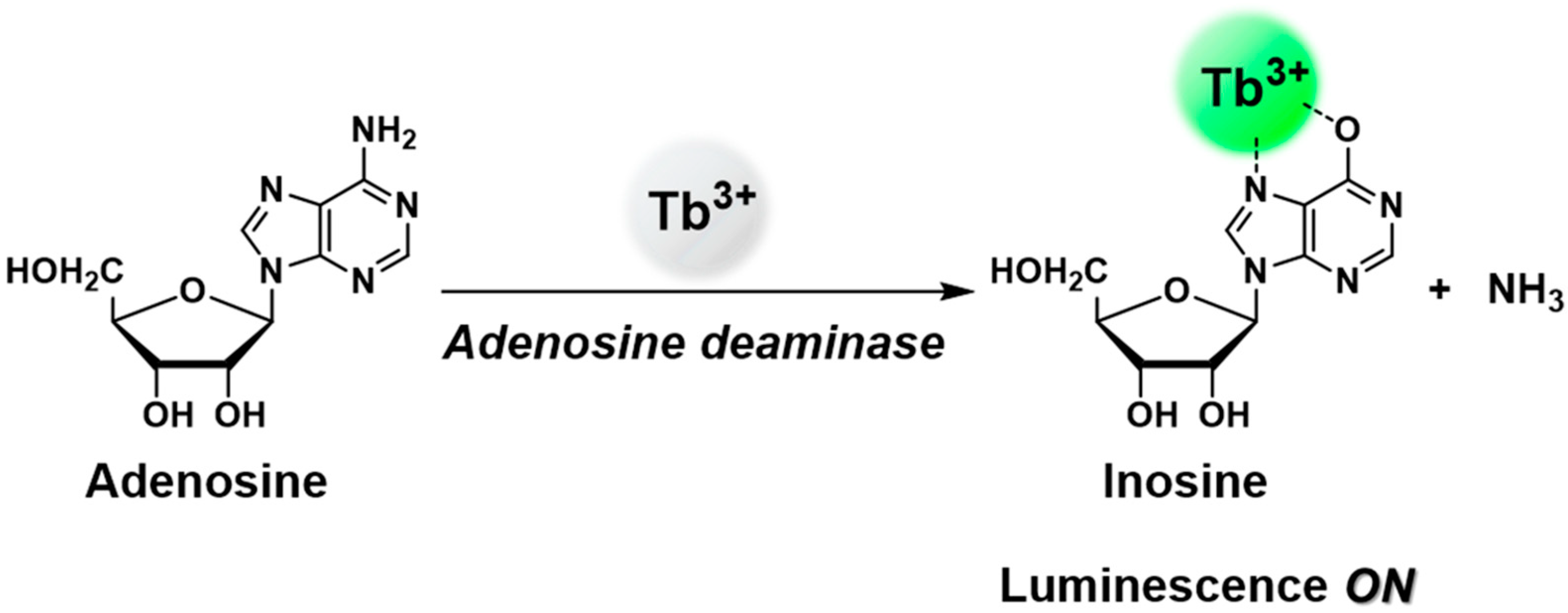
:1. Introduction
2. Materials and Methods
2.1. Materials and Instrumentation
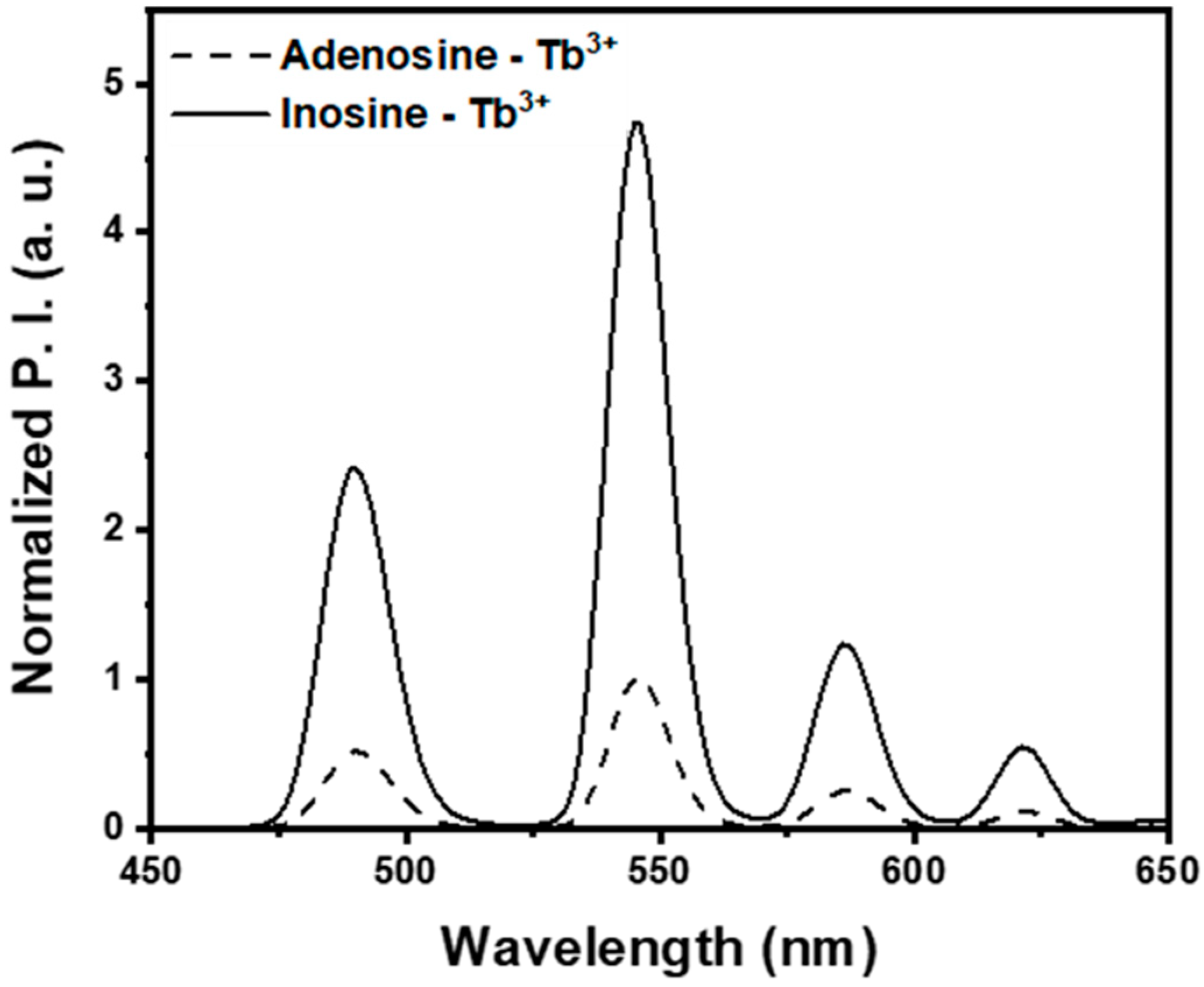
2.2. Optimization of Condition for Discrimination Between Adenosine and Inosine
2.3. Model study of Enzymatic Antenna Formation
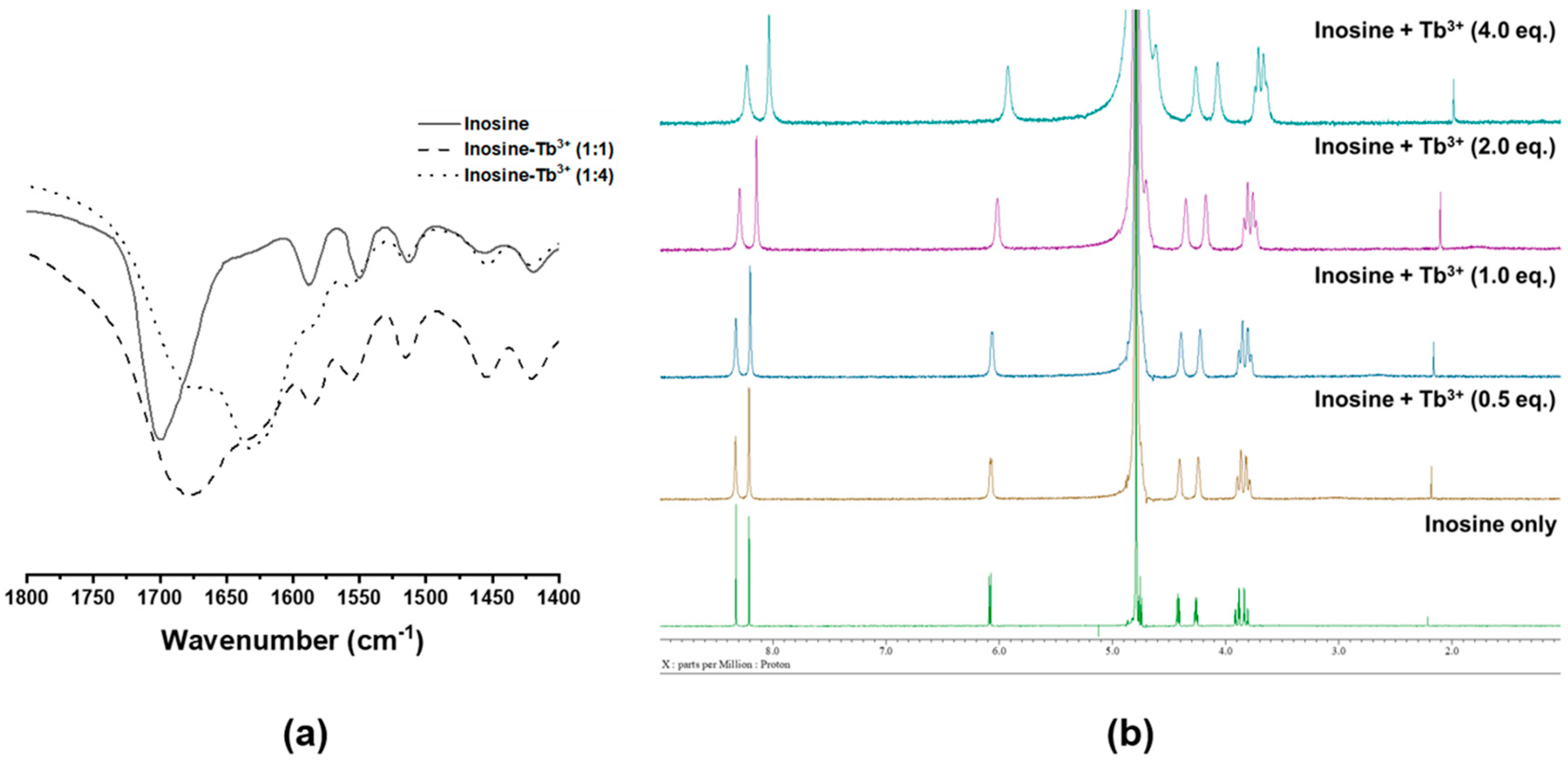
2.4. Identification of Chemical Structure of Inosine-Tb3+ Complex by FT-IR and 1H NMR Spectroscopy
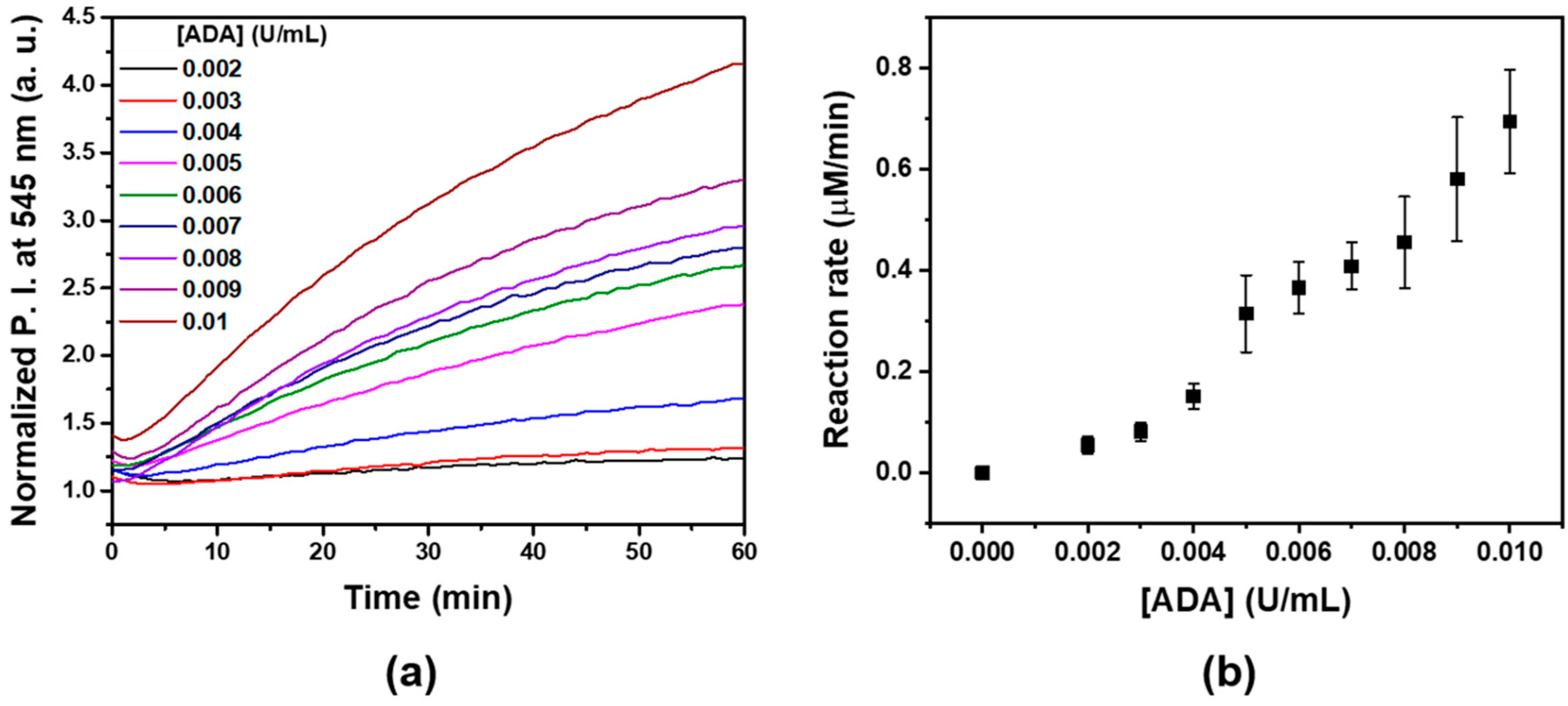
2.5. Kinetic Measurement of ADA Enzyme Reaction
2.6. Detection of ADA Inhibition by EHNA
3. Results and Discussion
3.1. Optimization of ADA Assay Condition Using Lanthanide Ion
3.2. Kinetic Measurement of ADA Enzyme Reaction
3.3. Determination of Inhibition Efficiency of ADA Inhibitor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Van der Weyden, M.B.; Kelley, W.N. Human Adenosine Deaminase. Distribution and properties. J. Biol. Chem. 1976, 251, 5448–5456. [Google Scholar]
- Cristalli, G.; Costanzi, S.; Lambertucci, C.; Lupidi, G.; Vittori, S.; Volpini, R.; Camaioni, E. Adenosine Deaminase: Functional Implications and Different Classes of Inhibitors. Med. Res. Rev. 2001, 21, 105–128. [Google Scholar] [CrossRef]
- Cohen, A.; Hirschhorn, R.; Horowitz, S.D.; Rubinstein, A.; Polmar, S.H.; Hong, R.; Martin, D.W., Jr. Deoxyadenosine triphosphate as a potentially toxic metabolite in adenosine deaminase deficiency. Proc. Natl. Acad. Sci. USA 1978, 75, 472–476. [Google Scholar] [CrossRef]
- Hirschhorn, R. Adenosine Deaminase Deficiency: Molecular Basis and Recent Developments. Clin. Immunol. Immunopathol. 1995, 76, S219–S227. [Google Scholar] [CrossRef]
- Agarwal, R.P.; Crabtree, G.W.; Parks, R.E., Jr.; Nelson, J.A.; Keightley, R.; Parkman, R.; Rosen, F.S.; Stern, R.C.; Polmar, S.H. Purine nucleoside metabolism in the erythrocytes of patients with adenosine deaminase deficiency and severe combined immunodeficiency. J. Clin. Investig. 1976, 57, 1025–1035. [Google Scholar] [CrossRef]
- Donofrio, J.; Coleman, M.S.; Hutton, J.J.; Daoud, A.; Lampkin, B.; Dyminski, J. Overproduction of adenine deoxynucleosides and deoxynucleotides in adenosine deaminase deficiency with severe combined immunodeficiency disease. J. Clin. Investig. 1978, 62, 884–887. [Google Scholar] [CrossRef]
- Coleman, M.S.; Donofrio, J.; Hutton, J.J.; Hahn, L. Identification and Quantitation of Adenine Deoxynucleotides in Erythrocytes of a Patient with Adenosine Deaminase Deficiency and Severe Combined Immunodeficiency. J. Biol. Chem. 1978, 253, 1619–1626. [Google Scholar]
- Aiuti, A.; Cattaneo, F.; Galimberti, S.; Benninghoff, U.; Cassani, B.; Callegaro, L.; Scaramuzza, S.; Andolfi, G.; Mirolo, M.M.; Brigida, I.; et al. Gene Therapy for Immunodeficiency Due to Adenosine Deaminase Deficiency. N. Engl. J. Med. 2009, 360, 447–458. [Google Scholar] [CrossRef]
- Smyth, J.F.; Poplack, D.G.; Holiman, B.J.; Leventhal, B.G. Correlation of adenosine deaminase activity with cell surface markers in acute lymphoblastic leukemia. J. Clin. Investig. 1978, 62, 710–712. [Google Scholar] [CrossRef]
- Hoffbrand, A.; Janossy, G. Enzyme and membrane markers in leukaemia: Recent developments. J. Clin. Pathol. 1981, 34, 254–262. [Google Scholar] [CrossRef]
- Segura, R.M.; Pascual, C.; Ocaña, I.; Martínez-Vázquez, J.M.; Ribera, E.; Ruiz, I.; Pelegrí, M.D. Adenosine Deaminase in Body Fluids: A Useful Diagnostic Tool in Tuberculosis. Clin. Biochem. 1989, 22, 141–148. [Google Scholar] [CrossRef]
- Burgess, L.J.; Swanepoel, C.G.; Taljaard, J.J.F. The use of adenosine deaminase as a diagnostic tool for peritoneal tuberculosis. Tuberculosis 2001, 81, 243–248. [Google Scholar] [CrossRef]
- Xia, Y.; Khatchikian, G.; Zweier, J.L. Adenosine Deaminase Inhibition Prevents Free Radical-mediated Injury in the Postischemic Heart. J. Biol. Chem. 1996, 271, 10096–10102. [Google Scholar] [CrossRef] [Green Version]
- Kutryb-Zajac, B.; Koszalka, P.; Mierzejewska, P.; Bulinska, A.; Zabielska, M.A.; Brodzik, K.; Skrzypkowska, A.; Zelazek, L.; Pelikant-Malecka, I.; Slominska, E.M.; et al. Adenosine deaminase inhibition suppresses progression of 4T1 murine breast cancer by adenosine receptor-dependent mechanisms. J. Cell. Mol. Med. 2018, 22, 5939–5954. [Google Scholar] [CrossRef]
- Hopkinson, D.A.; Cook, P.J.L.; Harris, H. Further data on the adenosine deaminase (ADA) polymorphism and a report of a new phenotype. Ann. Hum. Genet. 1969, 32, 361–367. [Google Scholar] [CrossRef]
- Oosthuizen, H.M.; Ungerer, J.P.J.; Bissbort, S.H. Kinetic Determination of Serum Adenosine Deaminase. Clin. Chem. 1993, 39, 2182–2185. [Google Scholar]
- Zhang, K.; Zhu, X.; Wang, J.; Xu, L.; Li, G. Strategy to Fabricate an Electrochemical Aptasensor: Application to the Assay of Adenosine Deaminase Activity. Anal. Chem. 2010, 82, 3207–3211. [Google Scholar] [CrossRef]
- Xing, X.J.; Liu, X.G.; Luo, Q.Y.; Tang, H.W.; Pang, D.W. Graphene oxide based fluorescent aptasensor for adenosine deaminase detection using adenosine as the substrate. Biosens. Bioelectron. 2012, 37, 61–67. [Google Scholar] [CrossRef]
- Zhang, K.; Xie, M.; Zhou, B.; Hua, Y.; Yan, Z.; Liu, H.; Guo, L.; Wu, B.; Huang, B. A new strategy based on aptasensor to time-resolved fluorescence assay for adenosine deaminase activity. Biosens. Bioelectron. 2013, 41, 123–128. [Google Scholar] [CrossRef]
- Wang, C.; Tang, Y.; Liu, Y.; Guo, Y. Water-Soluble Conjugated Polymer as a Platform for Adenosine Deaminase Sensing Based on Fluorescence Resonance Energy Transfer Technique. Anal. Chem. 2014, 86, 6433–6438. [Google Scholar] [CrossRef]
- Hu, K.; Huang, Y.; Wang, S.; Zhao, S. A carbon nanotubes based fluorescent aptasensor for highly sensitive detection of adenosine deaminase activity and inhibitor screening in natural extracts. J. Pharm. Biomed. Anal. 2014, 95, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; He, Y.; Xing, X.J.; Tan, D.D.; Lin, Y.; Pang, D.W.; Tang, H.W. A gold nanoparticle-based label free colorimetric aptasensor for adenosine deaminase detection and inhibition assay. Analyst 2015, 140, 1572–1577. [Google Scholar] [CrossRef]
- Bünzli, J.C.G.; Piguet, C. Taking advantage of luminescent lanthanide ions. Chem. Soc. Rev. 2005, 34, 1048–1077. [Google Scholar] [CrossRef] [PubMed]
- Massue, J.; Quinn, S.J.; Gunnlaugsson, T. Lanthanide Luminescent Displacement Assays: The Sensing of Phosphate Anions Using Eu (III)-Cyclen-Conjugated Gold Nanoparticles in Aqueous Solution. J. Am. Chem. Soc. 2008, 130, 6900–6901. [Google Scholar] [CrossRef] [PubMed]
- Mizukami, S.; Tonai, K.; Kaneko, M.; Kikuchi, K. Lanthanide-Based Protease Activity Sensors for Time-Resolved Fluorescence Measurements. J. Am. Chem. Soc. 2008, 130, 14376–14377. [Google Scholar] [CrossRef]
- Weitz, E.A.; Pierre, V.C. A ratiometric probe for the selective time-gated luminescence detection of potassium in water. Chem. Commun. 2011, 47, 541–543. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chang, H.; Xie, J.; Zhao, B.; Liu, B.; Xu, S.; Pei, W.; Ren, N.; Huang, L.; Huang, W. Recent developments in lanthanide-based luminescent probes. Coord. Chem. Rev. 2014, 273–274, 201–212. [Google Scholar] [CrossRef]
- Pershagen, E.; Borbas, K.E. Multiplex Detection of Enzymatic Activity with Responsive Lanthanide-Based Luminescent Probes. Angew. Chem. Int. Ed. 2015, 54, 1787–1790. [Google Scholar] [CrossRef]
- Aulsebrook, M.L.; Graham, B.; Grace, M.R.; Tuck, K.L. Lanthanide complexes for luminescence-based sensing of low molecular weight analytes. Coord. Chem. Rev. 2018, 375, 191–220. [Google Scholar] [CrossRef]
- Sabbatini, N.; Guardigli, M. Luminescent lanthanide complexes as photochemical supramolecular devices. Coord. Chem. Rev. 1993, 123, 201–228. [Google Scholar] [CrossRef]
- Moore, E.G.; Samuel, A.P.S.; Raymond, K.N. From Antenna to Assay: Lessons Learned in Lanthanide Luminescence. Accounts Chem. Res. 2009, 42, 542–552. [Google Scholar] [CrossRef] [Green Version]
- White, K.A.; Chengelis, D.A.; Gogick, K.A.; Stehman, J.; Rosi, N.L.; Petoud, S. Near-Infrared Luminescent Lanthanide MOF Barcodes. J. Am. Chem. Soc. 2009, 131, 18069–18071. [Google Scholar] [CrossRef] [PubMed]
- Armelao, L.; Quici, S.; Barigelletti, F.; Accorsi, G.; Bottaro, G.; Cavazzini, M.; Tondello, E. Design of luminescent lanthanide complexes: From molecules to highly efficient photo-emitting materials. Coord. Chem. Rev. 2010, 254, 487–505. [Google Scholar] [CrossRef]
- An, J.; Shade, C.M.; Chengelis-Czegan, D.A.; Petoud, S.; Rosi, N.L. Zinc-Adeninate Metal-Organic Framework for Aqueous Encapsulation and Sensitization of Near-infrared and Visible Emitting Lanthanide Cations. J. Am. Chem. Soc. 2011, 133, 1220–1223. [Google Scholar] [CrossRef] [PubMed]
- Georges, J. Lanthanide-sensitized Luminescence and Applications to the Determination of Organic Analytes. Analyst 1993, 118, 1481–1486. [Google Scholar] [CrossRef]
- Azab, H.A.; Al-Deyab, S.S.; Anwar, Z.M.; Abd El-Gawad, I.I.; Kamel, R.M. Comparison of the Coordination Tendency of Amino Acids, Nucleobases, or Mononucleotides Toward the Monomeric and Dimeric Lanthanide Complexes with Biologically Important Compounds. J. Chem. Eng. Data 2011, 56, 2613–2625. [Google Scholar] [CrossRef]
- Berezin, M.Y.; Achilefu, S. Fluorescence Lifetime Measurements and Biological Imaging. Chem. Rev. 2010, 110, 2641–2684. [Google Scholar] [CrossRef] [Green Version]
- Pneumatikakis, G.; Hadjiliadis, N.; Theophanides, T. Complexes of Inosine, Cytidine, and Guanosine with Palladium (II). Inorg. Chem. 1978, 17, 915–922. [Google Scholar] [CrossRef]
- Ma, C.; Tan, H.; Chen, L.; Song, Y.; Xu, F.; Chen, S.; Wang, L. A terbium chelate based fluorescent assay for alkaline phosphatase in biological fluid. Sens. Actuator B-Chem. 2014, 202, 683–689. [Google Scholar] [CrossRef]
- Wilmott, N.J.; Niller, J.N.; Tyson, J.F. Potential Use of a Terbium—Transferrin Complex as a Label in an Immunoassay for Gentamicin. Analyst 1984, 109, 343–345. [Google Scholar] [CrossRef]
- Ataie, G.; Bagheri, S.; Divsalar, A.; Saboury, A.A.; Safarian, S.; Namaki, S.; Moosavi-Movahedi, A.A. A Kinetic Comparison on the Inhibition of Adenosine Deaminase by Purine Drugs. Iran. J. Pharm. Res. 2007, 6, 43–50. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Park, H.; Ki, Y.; Lee, H.; Han, M.S. Development of a Simple Assay Method for Adenosine Deaminase via Enzymatic Formation of an Inosine-Tb3+ Complex. Sensors 2019, 19, 2728. https://doi.org/10.3390/s19122728
Lee S, Park H, Ki Y, Lee H, Han MS. Development of a Simple Assay Method for Adenosine Deaminase via Enzymatic Formation of an Inosine-Tb3+ Complex. Sensors. 2019; 19(12):2728. https://doi.org/10.3390/s19122728
Chicago/Turabian StyleLee, Suji, Heewon Park, Yeongcheol Ki, Hohjai Lee, and Min Su Han. 2019. "Development of a Simple Assay Method for Adenosine Deaminase via Enzymatic Formation of an Inosine-Tb3+ Complex" Sensors 19, no. 12: 2728. https://doi.org/10.3390/s19122728
APA StyleLee, S., Park, H., Ki, Y., Lee, H., & Han, M. S. (2019). Development of a Simple Assay Method for Adenosine Deaminase via Enzymatic Formation of an Inosine-Tb3+ Complex. Sensors, 19(12), 2728. https://doi.org/10.3390/s19122728





