Recognition of Pharmacological Bi-Heterocyclic Compounds by Using Terahertz Time Domain Spectroscopy and Chemometrics
Abstract
:1. Introduction
1.1. Test Methods Used to Diagnose Drugs
1.2. Drug Detection Using Terahertz Spectroscopy Combined with Support Vector Machine (Svm)
2. Experimental Study
2.1. TDS-THz Spectrometer
2.2. Method of Manufacture of the Samples
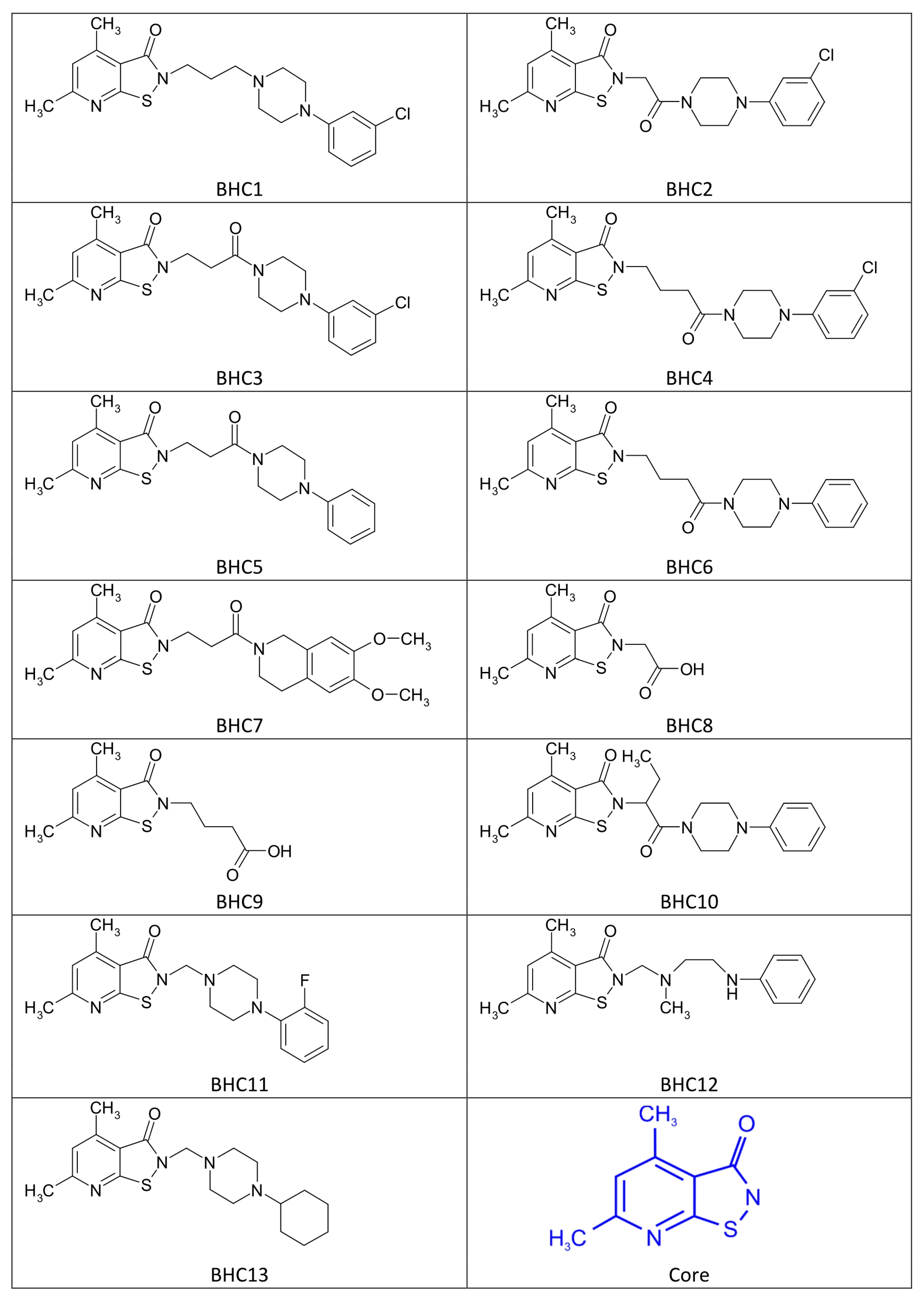
2.3. Chemical Structure of Bi-Heterocyclic Compounds
3. Statistical Analysis
3.1. PCA
3.2. KSVM
- d-degree polynomial:, where and ,
- Gaussian Radial Basis Functions (GRBF):, where
- two-layer perceptron:, where .
4. Classification Results
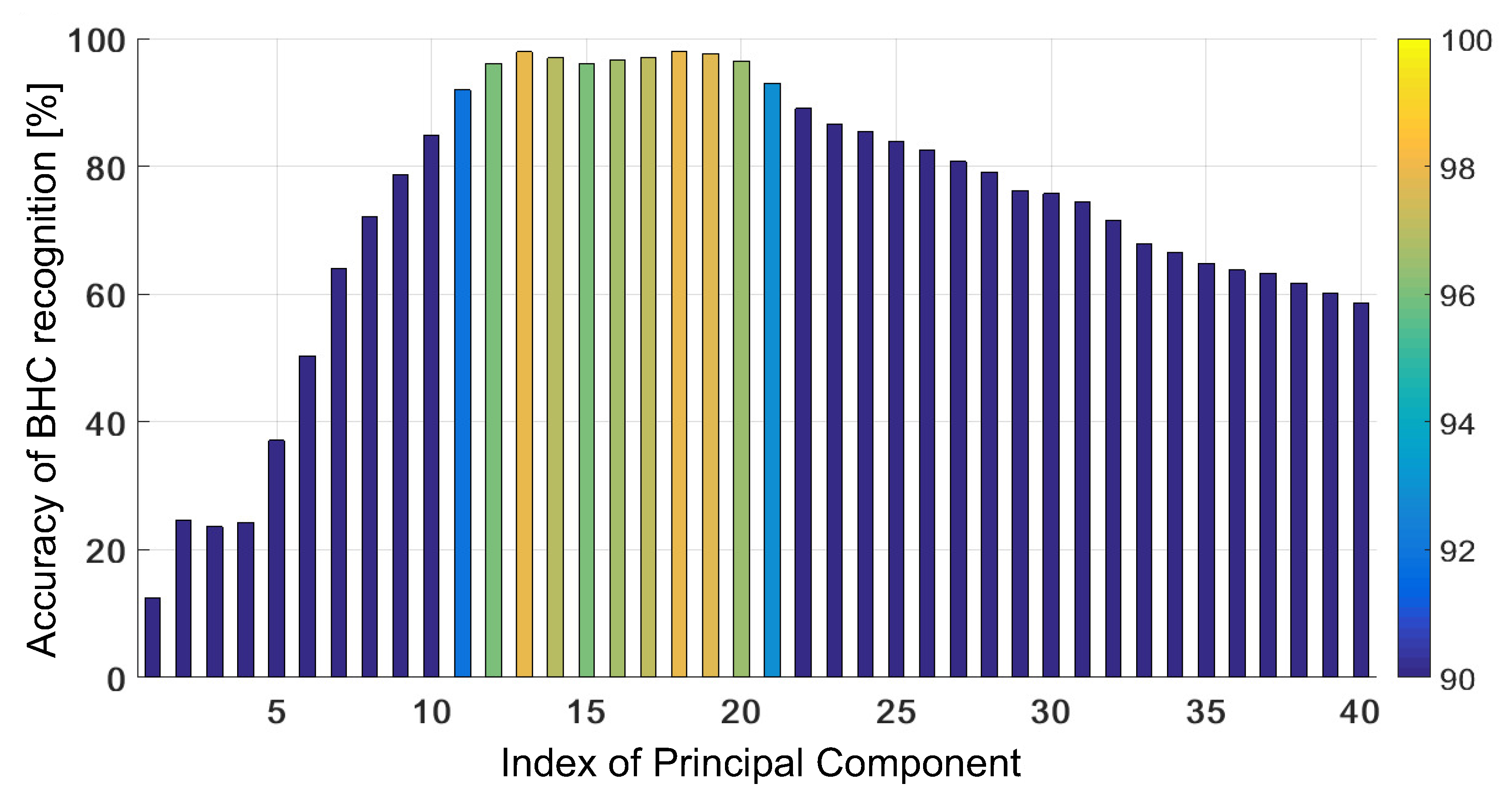
4.1. Untrained GRBF Kernel
4.2. Trained GRBF Kernel
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eicher, T.; Hauptmann, S.; Speicher, A. The Chemistry of Heterocycles: Structures, Reactions, Synthesis, and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Joule, J.A.; Mills, K. Heterocyclic Chemistry at a Glance; John Wiley & Sons: West Sussex, UK, 2012. [Google Scholar]
- Yanagimoto, K.; Lee, K.G.; Ochi, H.; Shibamoto, T. Antioxidative activity of heterocyclic compounds found in coffee volatiles produced by Maillard reaction. J. Agric. Food Chem. 2002, 50, 5480–5484. [Google Scholar] [CrossRef] [PubMed]
- Agranat, I. Pharmaceutical Substances: Syntheses, Patents and Applications of the Most Relevant AIPs. Synthesis 2009, 2009, 2116. [Google Scholar] [CrossRef]
- Lamberth, C.; Dinges, J. Bioactive Heterocyclic Compound Classes: Pharmaceuticals; Wiley-VCH: Weinheim, Germany, 2012. [Google Scholar]
- Kujawski, J.; Janusz, A.; Kuźma, W.; Ożarowski, M.; Kujawski, R. Biologicznie ważne substancje zawierające pierścień pirolu–działanie i występowanie. Farm Pol. 2010, 66, 117–125. [Google Scholar]
- Selvam, T.P.; James, C.R.; Dniandev, P.V.; Valzita, S.K. A mini review of pyrimidine and fused pyrimidine marketed drugs. Res. Pharm. 2012, 2, 4. [Google Scholar]
- Selvam, T.P.; Kumar, P.V. Quinazoline marketed drugs. Res. Pharm. 2011, 1, 1–21. [Google Scholar]
- Majumdar, K.C.; Chattopadhyay, S.K. Heterocycles in Natural Product Synthesis; Wiley-VCH: Weinheim, Germany, 2011. [Google Scholar]
- Alvarez-Builla, J.; Vaquero, J.J.; Barluenga, J. Modern Heterocyclic Chemistry, 4 Volume Set; Wiley-VCH: Weinheim, Germany, 2011; Volume 2. [Google Scholar]
- Paudel, A.; Raijada, D.; Rantanen, J. Raman spectroscopy in pharmaceutical product design. Adv. Drug Deliv. Rev. 2015, 89, 3–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, C.; Nyadong, L.; Yang, F.; Fernandez, F.M.; Brown, C.D.; Newton, P.N.; Kazarian, S.G. Assessment of hand-held Raman instrumentation for in situ screening for potentially counterfeit artesunate antimalarial tablets by FT-Raman spectroscopy and direct ionization mass spectrometry. Anal. Chim. Acta 2008, 623, 178–186. [Google Scholar] [CrossRef]
- Jamrógiewicz, M. Application of the near-infrared spectroscopy in the pharmaceutical technology. J. Pharm. Biomed. Anal. 2012, 66, 1–10. [Google Scholar] [CrossRef]
- Igne, B.; Ciurczak, E.W. Pharmaceutical and Medical Applications of Near-Infrared Spectroscopy; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- De Beer, T.; Burggraeve, A.; Fonteyne, M.; Saerens, L.; Remon, J.P.; Vervaet, C. Near infrared and Raman spectroscopy for the in-process monitoring of pharmaceutical production processes. Int. J. Pharm. 2011, 417, 32–47. [Google Scholar] [CrossRef] [Green Version]
- Bressolle, F.; Audran, M.; Pham, T.N.; Vallon, J.J. Cyclodextrins and enantiomeric separations of drugs by liquid chromatography and capillary electrophoresis: Basic principles and new developments. J. Chromatogr. Biomed. Sci. Appl. 1996, 687, 303–336. [Google Scholar] [CrossRef]
- Phadnis, N.V.; Cavatur, R.K.; Suryanarayanan, R. Identification of drugs in pharmaceutical dosage forms by X-ray powder diffractometry. J. Pharm. Biomed. Anal. 1997, 15, 929–943. [Google Scholar] [CrossRef]
- Rösner, P.; Junge, T.; Westphal, F.; Fritschi, G. Mass Spectra of Designer Drugs 2015; Wiley-VCH: Weinheim, Germany, 2015. [Google Scholar]
- Hummel, D.; Löffler, D.; Fink, G.; Ternes, T.A. Simultaneous determination of psychoactive drugs and their metabolites in aqueous matrices by liquid chromatography mass spectrometry. Environ. Sci. Technol. 2006, 40, 7321–7328. [Google Scholar] [CrossRef] [PubMed]
- Barras, J.; Althoefer, K.; Rowe, M.; Poplett, I.; Smith, J. The emerging field of medicines authentication by nuclear quadrupole resonance spectroscopy. Appl. Magn. Reson. 2012, 43, 511–529. [Google Scholar] [CrossRef]
- Barras, J.; Murnane, D.; Althoefer, K.; Assi, S.; Rowe, M.D.; Poplett, I.J.; Kyriakidou, G.; Smith, J.A. Nitrogen-14 nuclear quadrupole resonance spectroscopy: A promising analytical methodology for medicines authentication and counterfeit antimalarial analysis. Anal. Chem. 2013, 85, 2746–2753. [Google Scholar] [CrossRef] [PubMed]
- Buffler, A. Contraband detection with fast neutrons. Radiat. Phys. Chem. 2004, 71, 853–861. [Google Scholar] [CrossRef]
- Buffler, A.; Tickner, J. Detecting contraband using neutrons: Challenges and future directions. Radiat. Meas. 2010, 45, 1186–1192. [Google Scholar] [CrossRef]
- AlNabooda, M.O.; Shubair, R.M.; Rishani, N.R.; Aldabbagh, G. Terahertz spectroscopy and imaging for the detection and identification of illicit drugs. In Proceedings of the Sensors Networks Smart And Emerging Technologies (Senset), Beirut, Lebanon, 12–14 September 2017; pp. 1–4. [Google Scholar]
- Puc, U.; Abina, A.; Rutar, M.; Zidanšek, A.; Jeglič, A.; Valušis, G. Terahertz spectroscopic identification of explosive and drug simulants concealed by various hiding techniques. Appl. Opt. 2015, 54, 4495–4502. [Google Scholar] [CrossRef] [PubMed]
- Federici, J.F.; Schulkin, B.; Huang, F.; Gary, D.; Barat, R.; Oliveira, F.; Zimdars, D. THz imaging and sensing for security applications—Explosives, weapons and drugs. Semicond. Sci. Technol. 2005, 20, S266–S280. [Google Scholar] [CrossRef]
- Burnett, A.D.; Cunningham, J.E.; Davies, A.G.; Dean, P.; Linfield, E.H. Terahertz frequency spectroscopy and its potential for security applications. In Infrared and Raman Spectroscopy in Forensic Science; Wiley Online Library: West Sussex, UK, 2012; pp. 295–314. [Google Scholar]
- Kawase, K.; Ogawa, Y.; Watanabe, Y.; Inoue, H. Non-destructive terahertz imaging of illicit drugs using spectral fingerprints. Opt. Express 2003, 11, 2549–2554. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Tripathi, S.R.; Murate, K.; Imayama, K.; Kawase, K. Non-destructive drug inspection in covering materials using a terahertz spectral imaging system with injection-seeded terahertz parametric generation and detection. Opt. Express 2016, 24, 6425–6432. [Google Scholar] [CrossRef]
- Shen, Y.C.; Taday, P.F. Development and application of terahertz pulsed imaging for nondestructive inspection of pharmaceutical tablet. IEEE J. Sel. Top. Quantum Electron. 2008, 14, 407–415. [Google Scholar] [CrossRef]
- Ho, L.; Cuppok, Y.; Muschert, S.; Gordon, K.C.; Pepper, M.; Shen, Y.; Siepmann, F.; Siepmann, J.; Taday, P.F.; Rades, T. Effects of film coating thickness and drug layer uniformity on in vitro drug release from sustained-release coated pellets: A case study using terahertz pulsed imaging. Int. J. Pharm. 2009, 382, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Felton, L.A.; Porter, S.C. An update on pharmaceutical film coating for drug delivery. Expert Opin. Drug Deliv. 2013, 10, 421–435. [Google Scholar] [CrossRef] [PubMed]
- May, R.K.; Evans, M.J.; Zhong, S.; Warr, I.; Gladden, L.F.; Shen, Y.; Zeitler, J.A. Terahertz in-line sensor for direct coating thickness measurement of individual tablets during film coating in real-time. J. Pharm. Sci. 2011, 100, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Zeitler, J.A.; Shen, Y.C. Industrial applications of terahertz imaging. In Terahertz Spectroscopy and Imaging; Springer: Berlin/Heidelberg, Germany, 2013; pp. 451–489. [Google Scholar]
- Taday, P.F.; Bradley, I.; Arnone, D.; Pepper, M. Using terahertz pulse spectroscopy to study the crystalline structure of a drug: A case study of the polymorphs of ranitidine hydrochloride. J. Pharm. Sci. 2003, 92, 831–838. [Google Scholar] [CrossRef]
- Taday, P.F. Applications of terahertz spectroscopy to pharmaceutical sciences. Philos. Trans. R. Soc. Lond. Ser. Math. Phys. Eng. Sci. 2003, 362, 351–364. [Google Scholar] [CrossRef]
- Kojima, S.; Shibata, T.; Igawa, H.; Mori, T. Broadband terahertz time-domain spectroscopy: Crystalline and glassy drug materials. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Tsukuba, Japan, 2014; Volume 54, p. 012001. [Google Scholar]
- Du, S.; Li, H.; Xie, L.; Chen, L.; Peng, Y.; Zhu, Y.; Li, H.; Dong, P.; Wang, J. Vibrational frequencies of anti-diabetic drug studied by terahertz time-domain spectroscopy. Appl. Phys. Lett. 2012, 100, 143702. [Google Scholar] [CrossRef]
- Ueno, Y.; Ajito, K.; Kukutsu, N.; Tamechika, E. Quantitative analysis of amino acids in dietary supplements using terahertz time-domain spectroscopy. Anal. Sci. 2011, 27, 351. [Google Scholar] [CrossRef]
- Müller, J.; Brock, D.; Knop, K.; Zeitler, J.A.; Kleinebudde, P. Prediction of dissolution time and coating thickness of sustained release formulations using Raman spectroscopy and terahertz pulsed imaging. Eur. J. Pharm. Biopharm. 2012, 80, 690–697. [Google Scholar] [CrossRef]
- Shen, Y.C. Terahertz pulsed spectroscopy and imaging for pharmaceutical applications: A review. Int. J. Pharm. 2011, 417, 48–60. [Google Scholar] [CrossRef]
- Ergün, S.; Sönmez, S. Terahertz technology for military applications. J. Manag. Inf. Sci. 2015, 3, 13–16. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, X.; Yang, K.; Liu, Y.; Liu, Y.; Fu, W.; Luo, Y. Biomedical applications of terahertz spectroscopy and imaging. Trends Biotechnol. 2016, 34, 810–824. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; He, Y.; Liu, K.; Fan, S.; Parrott, E.P.; Pickwell-MacPherson, E. Recent advances in terahertz technology for biomedical applications. Quant. Imaging Med. Surg. 2017, 7, 345. [Google Scholar] [CrossRef] [PubMed]
- Song, H.J.; Nagatsuma, T. Handbook of Terahertz Technologies: Devices and Applications; CRC Press: Roca Raton, FL, USA, 2015. [Google Scholar]
- Saeedkia, D. Handbook of Terahertz Technology For Imaging, Sensing And Communications; Woodhead Publishing: Cambridge, UK, 2013. [Google Scholar]
- Dexheimer, S.L. Terahertz Spectroscopy: Principles and Applications; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Sterczewski, L.A.; Nowak, K.; Szlachetko, B.; Grzelczak, M.P.; Szczesniak-Siega, B.; Plinska, S.; Malinka, W.; Plinski, E.F. Chemometric evaluation of THz spectral similarity for the selection of early drug candidates. Sci. Rep. 2017, 7, 14583. [Google Scholar] [CrossRef] [PubMed]
- Byvatov, E.; Fechner, U.; Sadowski, J.; Schneider, G. Comparison of support vector machine and artificial neural network systems for drug/nondrug classification. J. Chem. Inf. Comput. Sci. 2003, 43, 1882–1889. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, S.; Zararsiz, G.; Goksuluk, D. Drug/nondrug classification using support vector machines with various feature selection strategies. Comput. Methods Programs Biomed. 2014, 117, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Burbidge, R.; Trotter, M.; Buxton, B.; Holden, S. Drug design by machine learning: Support vector machines for pharmaceutical data analysis. Comput. Chem. 2001, 26, 5–14. [Google Scholar] [CrossRef]
- Louis, B.; Agrawal, V.K.; Khadikar, P.V. Prediction of intrinsic solubility of generic drugs using MLR, ANN and SVM analyses. Eur. J. Med. Chem. 2010, 45, 4018–4025. [Google Scholar] [CrossRef]
- Sorich, M.J.; Miners, J.O.; McKinnon, R.A.; Winkler, D.A.; Burden, F.R.; Smith, P.A. Comparison of linear and nonlinear classification algorithms for the prediction of drug and chemical metabolism by human UDP-glucuronosyltransferase isoforms. J. Chem. Inf. Comput. Sci. 2003, 43, 2019–2024. [Google Scholar] [CrossRef]
- Liu, M.; Wu, Y.; Chen, Y.; Sun, J.; Zhao, Z.; Chen, X.W.; Matheny, M.E.; Xu, H. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J. Am. Med Inform. Assoc. 2012, 19, e28–e35. [Google Scholar] [CrossRef] [Green Version]
- Heikamp, K.; Bajorath, J. Support vector machines for drug discovery. Expert Opin. Drug Discov. 2014, 9, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Y.; Zhao, G.; Wang, W.; Li, F. Chinese traditional medicine recognition by support vector machine (SVM) terahertz spectrum. Guang Xue Guang Fen 2009, 29, 2346–2350. [Google Scholar]
- Nowak, M.; Nowak, K.; Grzelczak, M.; Szlachetko, B.; Sterczewski, L.; Plinski, E.; Swiatek, P.; Strzelecka, M.; Plinska, S.; Malinka, W. Machine learning applied to bi-heterocyclic drugs recognition. In Proceedings of the 2017 42nd International Conference on Infrared, Millimeter, and Terahertz Waves (IRMMW-THz), Cancún, Mexico, 27 August–1 September 2017; pp. 1–2. [Google Scholar]
- Selzer, P.M. Comprehensive Analysis of Parasite Biology: From Metabolism to Drug Discovery; Wiley-VCH: Weinheim, Germany, 2016. [Google Scholar]
- Shibany, K.A.; Tötemeyer, S.; Pratt, S.L.; Paine, S.W. Equine hepatocytes: Isolation, cryopreservation, and applications to in vitro drug metabolism studies. Pharmacol. Res. Perspect. 2016, 4, e00268. [Google Scholar] [CrossRef] [PubMed]
- Platt, J. Sequential Minimal Optimization: A Fast Algorithm for Training Support Vector Machines. Technical Report MSR-TR-98-14, Microsoft Research. Available online: https://pdfs.semanticscholar.org/59ee/e096b49d66f39891eb88a6c84cc89acba12d.pdf (accessed on 28 June 2019).
- Zdunek, R.; Nowak, M.; Plinski, E. Statistical classification of soft solder alloys by laser-induced breakdown spectroscopy: Review of methods. J. Eur. Opt. Soc. Rapid Publ. 2016, 11, 16006. [Google Scholar] [CrossRef]
- Wojcik, M.R.; Zdunek, R.; Antonczak, A.J. Unsupervised verification of laser-induced breakdown spectroscopy dataset clustering. Spectrochim. Acta Part At. Spectrosc. 2016, 126, 84–92. [Google Scholar] [CrossRef]
- Hofmann, T.; Schölkopf, B.; Smola, A. Kernel Methods in Machine Learning. Ann. Stat. 2008, 36, 1171–1220. [Google Scholar] [CrossRef]
- Liu, W.; Principe, J.C.; Haykin, S. Kernel Adaptive Filtering: A Comprehensive Introduction, 1st ed.; Wiley Publishing: Hoboken, NJ, USA, 2010. [Google Scholar]
- Gendreau, M.; Potvin, J.Y. Handbook of Metaheuristics, 2nd ed.; Springer Publishing Company, Incorporated: New York, NY, USA, 2010. [Google Scholar]
- De Jong, K.A. Evolutionary Computation: A Unified Approach; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Lewis, R.M.; Torczon, V.; Trosset, M.W. Direct search methods: Then and now. J. Comput. Appl. Math. 2000, 124, 191–207. [Google Scholar] [CrossRef]
- Nelder, J.A.; Mead, R. A Simplex Method for Function Minimization. Comput. J. 1965, 7, 308–313. [Google Scholar] [CrossRef]







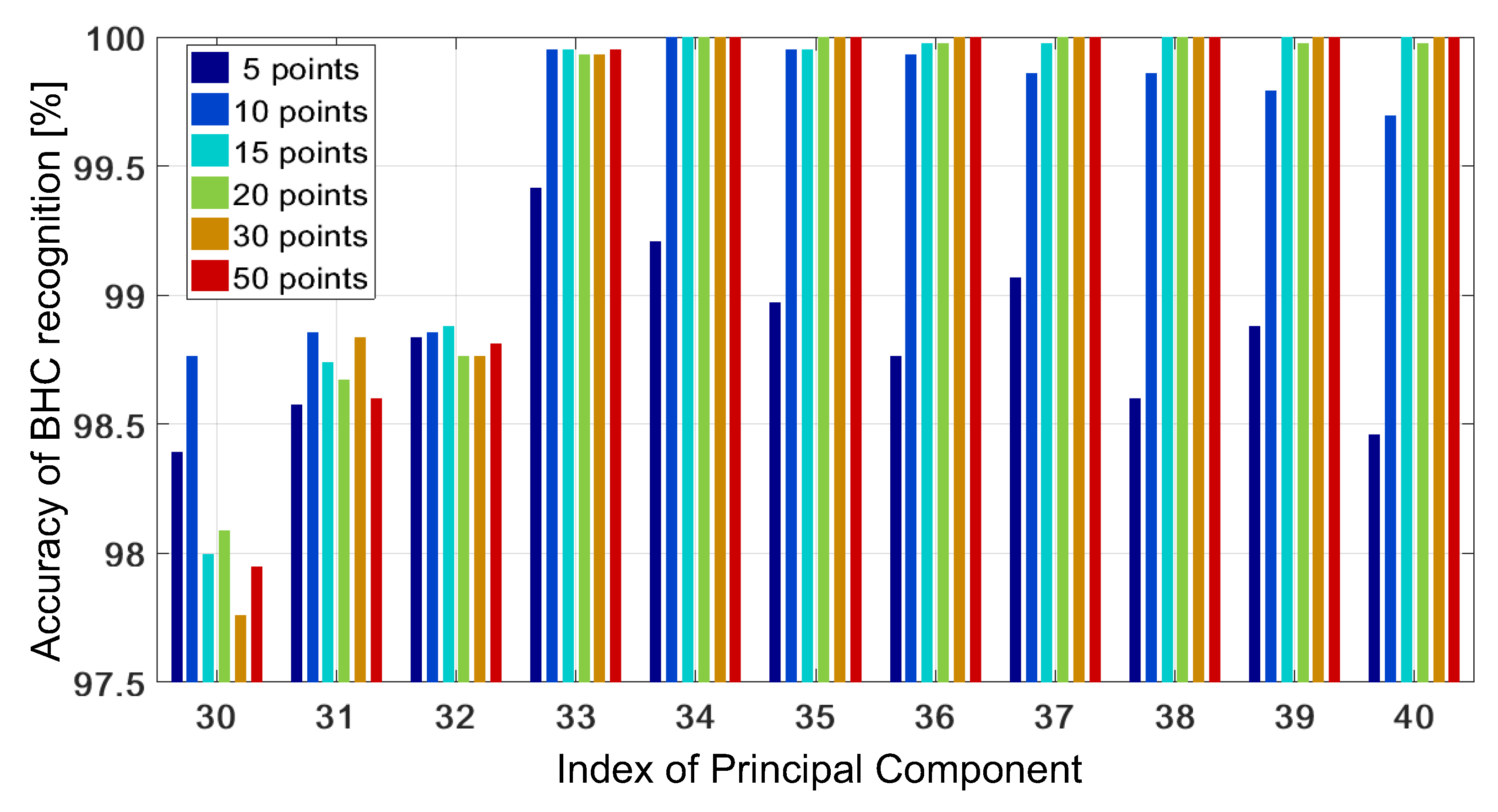
| No. initialization points: | 5 | 10 | 15 | 20 | 30 | 50 |
| Mean training time [s]: | 4.01 | 4.84 | 5.34 | 5.97 | 6.7 | 8.22 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowak, M.R.; Zdunek, R.; Pliński, E.; Świątek, P.; Strzelecka, M.; Malinka, W.; Plińska, S. Recognition of Pharmacological Bi-Heterocyclic Compounds by Using Terahertz Time Domain Spectroscopy and Chemometrics. Sensors 2019, 19, 3349. https://doi.org/10.3390/s19153349
Nowak MR, Zdunek R, Pliński E, Świątek P, Strzelecka M, Malinka W, Plińska S. Recognition of Pharmacological Bi-Heterocyclic Compounds by Using Terahertz Time Domain Spectroscopy and Chemometrics. Sensors. 2019; 19(15):3349. https://doi.org/10.3390/s19153349
Chicago/Turabian StyleNowak, Maciej Roman, Rafał Zdunek, Edward Pliński, Piotr Świątek, Małgorzata Strzelecka, Wiesław Malinka, and Stanisława Plińska. 2019. "Recognition of Pharmacological Bi-Heterocyclic Compounds by Using Terahertz Time Domain Spectroscopy and Chemometrics" Sensors 19, no. 15: 3349. https://doi.org/10.3390/s19153349






