1. Introduction
Conventional machine learning approaches are built upon a set of carefully engineered representations, which consist of measurable parameters extracted from raw data. Based on some expert knowledge in the domain of application, a feature extractor is designed and used to extract relevant information in the form of a feature vector from the preprocessed raw data. This high level representation of the input data is subsequently used to optimise an inference model. Although such approaches have proven to be very effective and can potentially lead to state-of-the-art results (given that the set of extracted descriptors is suitable for the task at hand), the corresponding performance and generalisation capability is limited by the reliance on expert knowledge as well as the inability of the designed model to process raw data directly and to dynamically adapt to related new tasks.
Meanwhile, deep learning approaches [
1] automatically generate suitable representations by applying a succession of simple and non-linear transformations on the raw data. A deep learning architecture consists of a hierarchical construct of several processing layers. Each processing layer is characterised by a set of parameters that are used to transform its input (which is the representation generated by the previous layer) into a new and more abstract representation. This specific hierarchical combination of several non-linear transformations enables deep learning architectures to learn very complex functions as well as abstract descriptive (or discriminative) representations directly from raw data [
2]. Moreover, the hierarchical construct characterising deep learning architectures offers more flexibility when it comes to adapting such approaches to new and related tasks. Hence, deep learning approaches have been outperforming previous state-of-the-art machine learning approaches, especially in the field of image processing [
3,
4,
5,
6,
7]. Similar performances have been achieved in the field of speech recognition [
8,
9] and natural language processing [
10,
11].
A steadily growing amount of work has been exploring the application of deep learning approaches on physiological signals. Martinéz et al. [
12] were able to significantly outperform standard approaches built upon hand-crafted features by using a deep learning algorithm for affect modelling based on physiological signals (two physiological signals consisting of Skin Conductance (SC) and Blood Volume Pulse (BVP) were used in this specific work). The designed approach consisted of a multi-layer Convolutional Neural Network (CNN) [
13] combined with a single-layer perceptron (SLP). The parameters of the CNN were trained in an unsupervised manner using denoising auto-encoders [
14]. The SLP was subsequently trained in a supervised manner using backpropagation [
15] to map the outputs of the CNN to the target affective states. In [
16], the authors proposed a multiple-fusion-layer based ensemble classifier of stacked auto-encoder (MESAE) for emotion recognition based on physiological data. A physiological-data-driven approach was proposed in order to identify the structure of the ensemble. The architecture was able to significantly outperform the existing state-of-the-art performance. A deep CNN was also successfully applied in [
17] for arousal and valence classification based on both electrocardiogram (ECG) and Galvanic Skin Response (GSR) signals. In [
18], a hybrid approach using CNN and Long Short-Term Memory (LSTM) [
19] Recurrent Neural Network (RNN) was designed to automatically extract and merge relevant information from several data streams stemming from different modalities (physiological signals, environmental and location data) for emotion classification. Moreover, deep learning approaches have been applied on electromyogram (EMG) signals for gesture recognition [
20,
21] or hand movement classification [
22,
23]. Most of the reported approaches consist of first transforming the processed EMG signal into a two dimensional (time-frequency) visual representation (such as a spectrogram or a scalogram) and subsequently using a deep CNN architecture to proceed with the classification. A similar procedure was used in [
24] for the analysis of electroencephalogram (EEG) signals. These are just some examples of an increasingly growing field of experimentation for deep neural networks. A better overview of deep learning approaches applied to physiological signals can be found in [
25,
26]. However, there are few related works that focus specifically on the application of deep neural networks on physiological signals for pain recognition. The authors of [
27] recently proposed a classification architecture based on Deep Belief Networks (DBNs) for the assessment of patients’ pain level during surgery, using photoplethysmography (PPG). The proposed architecture consists of a bagged ensemble of DBNs, built upon a set of manually engineered features, extracted from the recorded and preprocessed PPG signals. It is important to note that, in this specific study, the ensemble of bagged DBNs was trained on a set of carefully designed hand-crafted features. Therefore, an expert knowledge in this specific area of application is still needed in order to generate a set of relevant descriptors, since the whole classification process is not performed in an end-to-end manner.
Nonetheless, there is a constantly growing amount of works that focus specifically on pain recognition based on physiological signals, and categorised by the nature of the pain elicitations. There is a huge variety of statistical methods that have been proposed, most of them based on more traditional machine learning approaches such as decision trees or Support Vector Machines (SVMs) [
28]. In [
29], the authors proposed a continuous pain monitoring method using an Artificial Neural Network (ANN), based on hand-crafted features (wavelength (WL) and root mean square (RMS) features) extracted from several physiological signals consisting of heart rate (HR), breath rate (BR), galvanic skin response (GSR) and facial surface electromyogram (sEMG). The proposed approach was assessed on a dataset collected by inducing both thermal and electrical pain stimuli. In [
30], the authors proposed a pain detection approach based on EEG signals. Relevant features are extracted from the EEG signals using the Choi–Williams quadratic time–frequency distribution and subsequently used to train a SVM in order to perform the classification task. Pain in this specific work is elicited throughout tonic cold. Most recently, Thiam et al. [
31,
32] provided the results for a row of pain intensity classification experiments based on the
SenseEmotion Database (SEDB) [
33], by using several fusion architectures to merge hand-crafted features extracted from different modalities, including physiological, audio and video channels. Thereby, the combination of the features extracted from the recorded signals was compared for different fusion approaches, namely the fusion at feature level, the fusion at the classifiers’ output level and the fusion at an intermediate level. Random Forests [
34] were used as the base classifiers. In [
35], the authors combined camera PPG input signals with ECG and EMG signals in order to proceed with a user-independent pain intensity classification using the same dataset. The authors used a fusion architecture at the feature level with Random Forests and SVMs as base classifiers.
In [
36,
37,
38], the authors performed different pain intensity classification experiments based on the
BioVid Heat Pain Database [
39] (BVDB). All the conducted experiments were based on a carefully selected set of features extracted from both physiological and video channels. The classification was also performed using either Random Forests or SVMs. In [
40], Kächele et al. performed a user-independent pain intensity classification evaluation based on physiological input signals, using the same dataset. The authors used the whole data from all recorded pain levels in a classification, as well as a regression setting with Random Forests as the base classifiers. Several personalisation techniques were designed and validated, based on meta information from the test subjects, distance measures and machine learning techniques. The same authors proposed an adaptive confidence learning approach for personalised pain estimation in [
41] based on both physiological and video modalities. Thereby, the authors applied the fusion at feature level. The whole pain intensity estimation task was analysed as a regression problem. Random Forests were used as the base regression models. Moreover, a multi-layer perceptron (MLP) was applied to compute the confidence for an additional personalisation step. One recent work included the physiological signals of both datasets (SEDB and BVDB) [
42]. The authors analysed different fusion approaches with fixed aggregating rules based on their merging level for the person-independent multi-class scenario using all available pain levels. Thereby, three of the most popular decision tree based classifier systems, i.e., Bagging [
43], Boosting [
44] and Random Forests, were compared.
The current work focuses on the application of deep learning approaches for nociceptive heat-induced pain recognition based on physiological signals (EMG, ECG and electrodermal activity (EDA)). Several deep learning architectures are proposed for the assessment of measurable physiological parameters in order to perform an end-to-end classification of different levels of artificially induced nociceptive pain. The current work aims at achieving state-of-the-art classification performances based on feature learning (the designed architecture autonomously extracts relevant features from the preprocessed raw signals in an end-to-end manner), therefore removing the reliance on expert knowledge for the design and optimisation of reliable pain intensity classification models (since most of the previous works on pain intensity classification involving autonomic parameters rely on a carefully designed set of hand-crafted features). The remainder of the work is organised as follows. The proposed deep learning approaches as well as the dataset used for the validation of the approaches are described in
Section 2. Subsequently, a description of the results corresponding to the conducted assessments specific to each presented approach is provided in
Section 3. Finally, the findings of the conducted experiments are discussed in
Section 4, followed by the description of potential future works and a conclusion.
3. Results
All previously described deep architectures are trained using the Adaptive Moment estimation (
Adam) [
51] optimisation algorithm with a fixed learning rate set empirically to
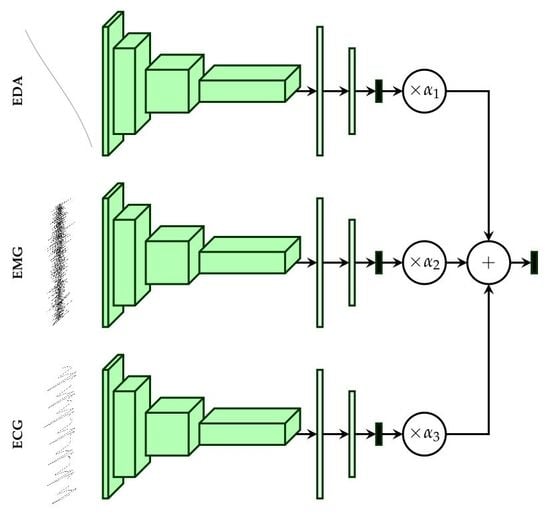
. The training process consisted of 100 epochs with the batch size set to 100. The weights of the loss function for the second late fusion architecture (see
Figure 4b) were empirically set as follows:
,
. The weight corresponding to the aggregation layer (
) was set higher than the others to push the network to focus on the weighted combination of the single modality architectures’ outputs, and therefore to evaluate an optimal set of the weighting parameters
. The implementation and evaluation of the described algorithms was done with the libraries Keras [
52], Tensorflow [
53] and Scikit-learn [
54]. The evaluation of the architectures was performed in a
Leave-One-Subject-Out (LOSO) cross-validation setting, which means that 87 experiments were conducted. During each experiment, the data specific to a single participant were used to evaluate the performance of the trained deep model and were never seen during the optimisation of this specific deep model. The data specific to each single participant were therefore used once as an unseen test set, and the results depicted in this section consist of averaged performance metrics from a set of 87 performance values.
A performance evaluation of the designed architectures in a binary classification task consisting of the discrimination between the baseline temperature
and the pain tolerance temperature
is reported in
Table 3. The achieved results based on CNNs are also compared to the state-of-the-art results reported in previous works. At a glance, the designed deep learning architectures outperform the state-of-the-art results in every setting, except for the ECG modality. Regarding the aggregation of all physiological modalities, the second late fusion architecture performs best and sets a new state-of-the-art fusion performance with an average accuracy of 84.40%, which even outperforms the best fusion results reported in [
41], where the authors could achieve an average classification performance of 83.1% by using both physiological and video features.
The deep architecture based on the EDA modality significantly outperforms all previously reported classification results with an average accuracy of 84.57%.
Based on these findings, further classification experiments were conducted, based on each physiological modality and also the best performing fusion architecture (Late Fusion (b)). The performance evaluation of the conducted experiments consisting of several binary classification experiments and a multi-class classification experiment is summarised in
Table 4.
EDA significantly outperforms both EMG and ECG in all conducted classification experiments and constitutes the best performing single modality, which is consistent with the results reported in previous works. Both EMG and ECG depict similar classification performances and also perform poorly for almost all classification experiments. The discrimination between the baseline temperature
and the pain threshold temperature
, as well as the two intermediate temperatures
and
, constitute very difficult classification experiments that both modalities are unable to perform successfully. However, the classification performances of both modalities for the classification tasks
vs.
and
vs.
are significantly above chance level, which shows that higher temperatures of elicitation cause observable and measurable responses in the recorded physiological signals, that can be used to perform the classification tasks at a certain degree of satisfaction. However, the overall performance of the fusion architecture is greatly affected by the significantly poor performance of both ECG and EMG in comparison to EDA. As can be seen in
Table 4, the EDA classification architecture outperforms the fusion architecture in almost all classification experiments (but not significantly), except for the classification task
vs.
and the multi-class classification task (the performance improvement of the fusion architecture is however not significant).
The information stemming from both modalities EMG and ECG harms the optimisation process of the fusion architecture due to its inconsistency. However, it can be seen in
Figure 5 that the fusion architecture is able to detect the sources of inconsistent information and dynamically adapt by systematically assigning higher weight values to EDA, while both ECG and EMG are assigned significantly lower weight values for all conducted classification tasks, and therefore improving the generalisation ability of the fusion architecture.
Subsequently, the performance of both EDA and late fusion architectures were further evaluated using different performance measures. In the case of binary classification experiments,
true positives(
tp) correspond to the number of correct acceptances,
false positives(
fp) correspond to the number of false acceptances,
true negatives(
tn) correspond to the number of correct rejections and
false negatives(
fn) correspond to the number of false rejections. These four values stem from the confusion matrix of an evaluated inference model and are used to define different performance measures. Those used for the current evaluation of the designed classification architectures are defined in
Table 5.
The performance evaluation of the EDA architecture is depicted in
Figure 6, while the performance evaluation of the fusion architecture is depicted in
Figure 7. Considering binary classification experiments, both architectures are able to consistently discriminate between the baseline temperature
and the other temperatures of pain elicitation. However, the performance of both architectures with regards to the five-class classification experiment suggests that the discrimination between all five levels of pain elicitation is a very challenging classification task. While the overall accuracy of each architecture is significantly above random performance (which is 20% in the case of a five-class classification task), the discrimination of the intermediate levels of pain elicitation remains very difficult, as can be seen in
Figure 8. Both baseline and pain tolerance temperatures
and
can be classified with a relatively good performance. The classification performance of
is barely above random performance and both
and
are mostly confused with
and
, respectively. These results are however consistent with previous works on the same dataset.
We therefore compared the performance of the EDA and proposed late fusion approach to early works. For the sake of fairness, we considered the related works performed on the exact same dataset, using the exact same evaluation settings (LOSO with all 87 participants). The results depicted in
Table 6 clearly show that the designed CNN architecture specific to EDA is able to consistently and significantly outperform previous approaches in all binary classification settings. Moreover, the authors of [
56,
57] reported overall accuracy performances of, respectively, 74.40% and 81.30% for the binary classification task
vs.
based uniquely on EDA. These approaches are also based on carefully designed hand-crafted features and are also significantly outperformed by the proposed CNN architecture specific to EDA.
Furthermore, we also compared the proposed late fusion approach with other fusion approaches proposed in early works. The results depicted in
Table 7 show that the proposed fusion approach outperforms previous approaches for the binary classification task
vs.
. Concerning the multi-class classification task, the proposed fusion approach also outperforms early approaches with an overall accuracy of 36.54%. The authors of [
41] reported an overall accuracy of 33% with a classification model based on physiological modalities, while Werner et al. [
58] reported an overall accuracy of 30.8% with a classification model based on head pose and facial activity descriptors.
Moreover, the designed fusion architecture was tested on the
BioVid Heat Pain Database (Part B). The database was generated using the same exact procedure as
Part A. However, it consists of 86 participants and two additional EMG signals (from the corrugator and the zygomaticus muscles) were recorded. In this evaluation, we used the same signals as in
Part A (EMG of the trapezius muscle, ECG and EDA), and used the same fusion architecture (Late fusion (b) depicted in
Figure 4b). The computed results were subsequently compared with those of previous works. The corresponding results are depicted in
Table 8.
The methods reported in previous works consist of fusion approaches involving all the recorded signals and based on hand-crafted features [
37,
56]. Although the fusion approach proposed in the current work (late fusion (b)) is based only on three of the recorded physiological signals, it is still able to outperform the previously proposed approaches, as depicted in
Table 8. Therefore, it is believed that the performance of the architecture can be further improved by including the remaining signals (EMG corrugator, EMG zygomaticus, and Video) in the proposed architecture.
4. Discussion and Conclusions
This work explored the application of deep neural networks for pain intensity classification based on physiological data including ECG, EMG and EDA. Several CNN architectures, based on 1D and 2D convolutional layers, were designed and assessed based on the BioVid Heat Pain Database (Part A). Furthermore, several deep fusion architectures were also proposed for the aggregation of relevant information stemming from all involved physiological modalities. The proposed architecture specific to EDA significantly outperformed the results presented in previous works in all classification settings. For the classification task vs. , EDA achieved a state-of-the-art average accuracy of 84.57%. The proposed late fusion approach based on a weighted average of each modality specific model’s output also achieved state-of-the-art performances (average accuracy of 84.40% for the classification task vs. ), but was unable to significantly outperform the deep model based uniquely on EDA.
Moreover, all designed architectures were trained in an
end-to-end manner. Therefore, it is believed that the pre-training and fine tuning at different levels of abstraction of the CNN architectures, as well as the combination with recurrent neural networks (in order to include the temporal aspect of the physiological signals in the inference model), could potentially improve the performance of the current system, since such approaches have been successfully applied in other domains of application such as facial expression recognition [
59,
60,
61]. Finally, the recorded video data provide an additional channel that can be integrated into the fusion architecture in order to improve the performance of the whole system. Therefore, the video modality should also be evaluated and assessed in combination with the physiological modalities.
In summary, the performed assessment suggests that deep learning approaches are relevant for the inference of pain intensity based on 1D physiological data, and such methods are able to significantly outperform traditional approaches based on hand-crafted features. Domain expert knowledge could be bypassed by enabling the designed deep architecture to learn relevant features from the data. In the future iterations of the current work, approaches consisting of combining both learned and hand-crafted features should be addressed. In addition, the designed architectures should be also assessed by replacing the classification experiments by regression experiments. Additionally, several data transformation approaches applied to the recorded 1D physiological data in order to generate 2D visual representations (e.g., spectrograms) should also be investigated in combination with established deep neural network approaches, specifically designed for this type of data representation.