Review of Electrochemical DNA Biosensors for Detecting Food Borne Pathogens
Abstract
1. Introduction
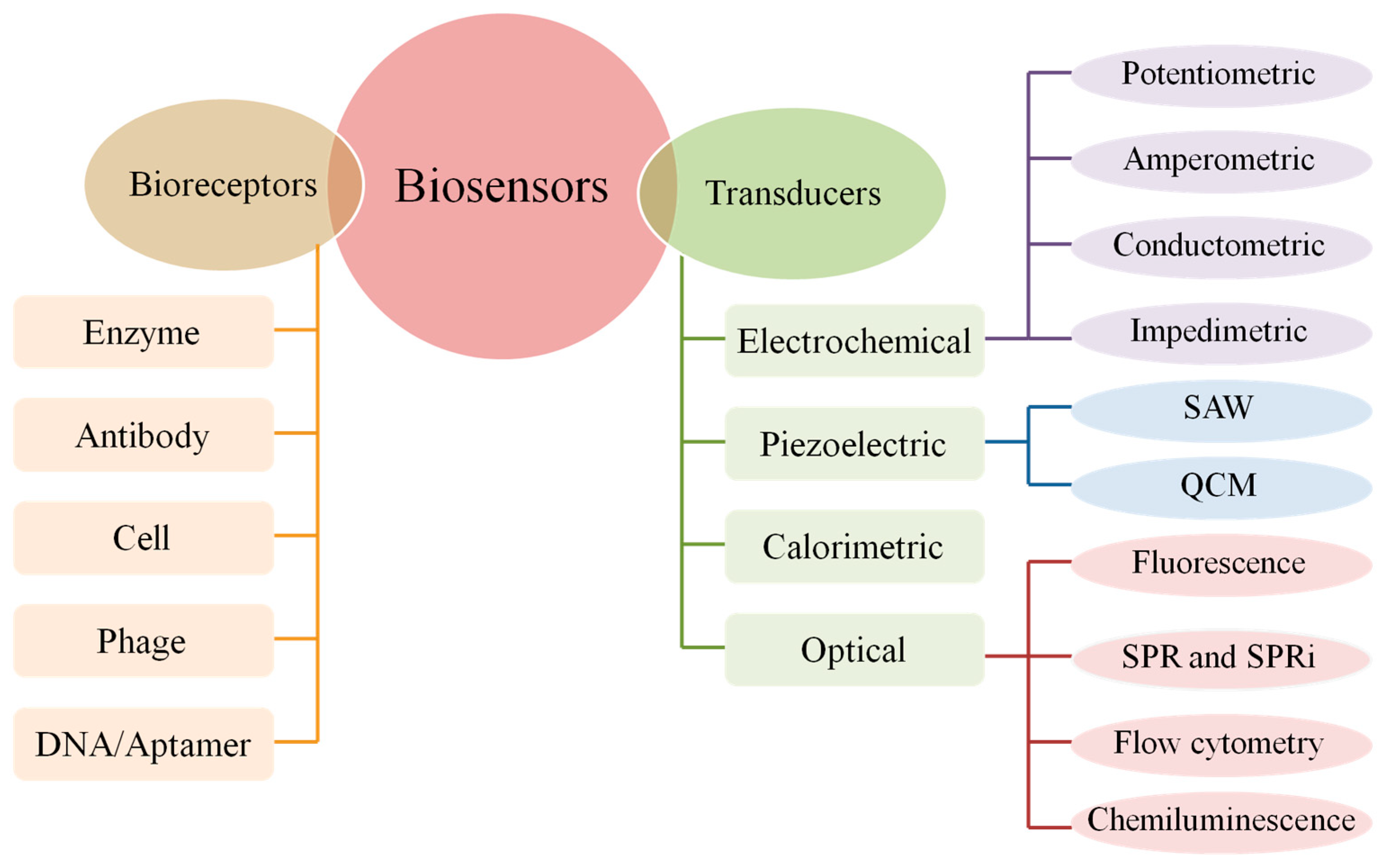
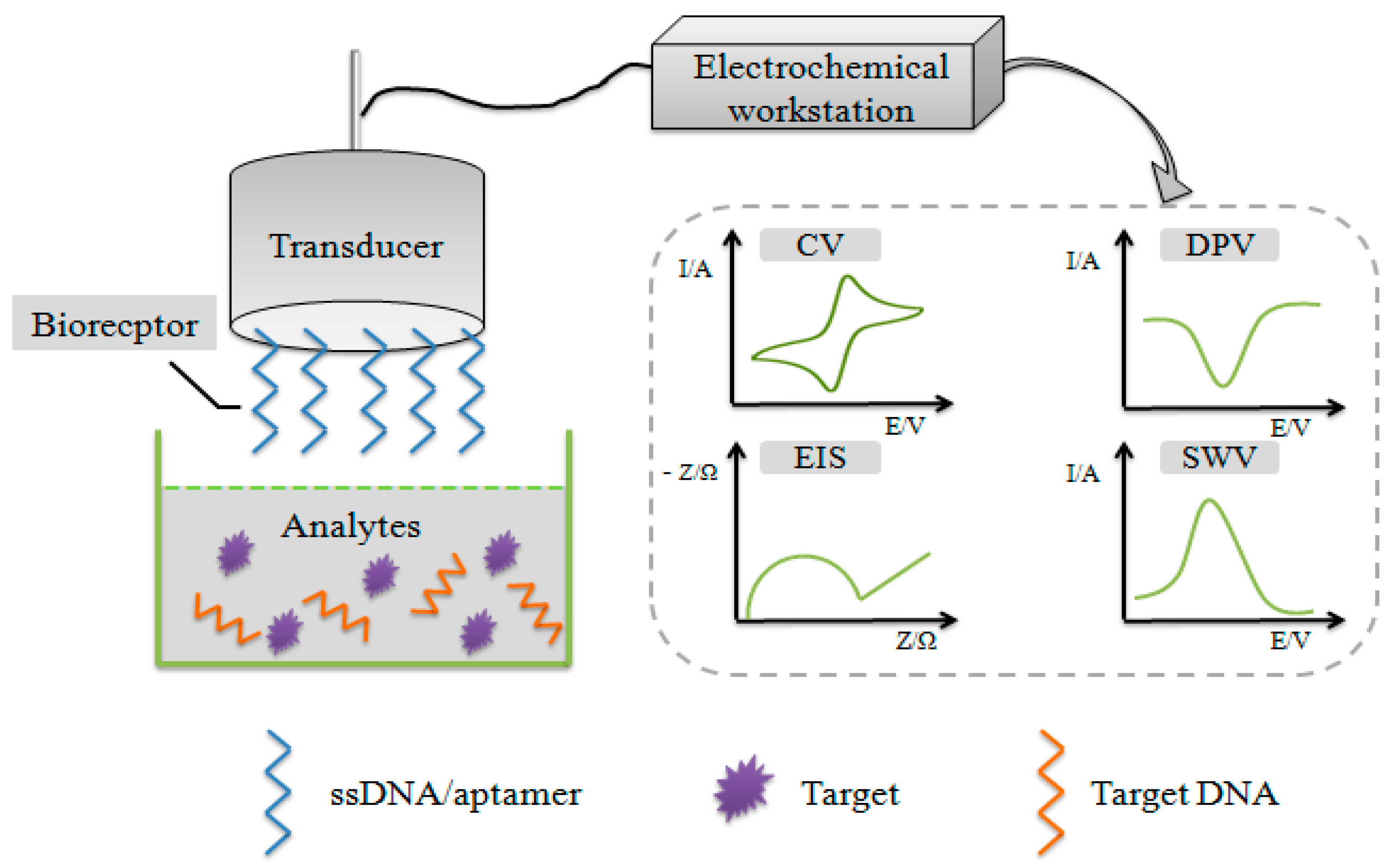
2. Electrochemical DNA Biosensors
2.1. Basic Principle of Electrochemical DNA Biosensors
2.2. Bioreceptor of Electrochemical DNA Biosensor
2.2.1. Type of Bioreceptor
Advantages of Aptamer
Detection Mechanisms of Aptamers with their Targets
2.2.2. Immobilization Methods of Bioreceptor
2.3. Electrochemical Techniques
2.4. Detection Methods
3. Strategies for Improving the Sensitivity of Electrochemical DNA Biosensors
3.1. Nanomaterials
3.1.1. Conventional Nanomaterials
3.1.2. Composite Nanomaterial
3.1.3. Emerging Nanomaterials
3.2. Nucleic Acid-Based Amplification Technologies
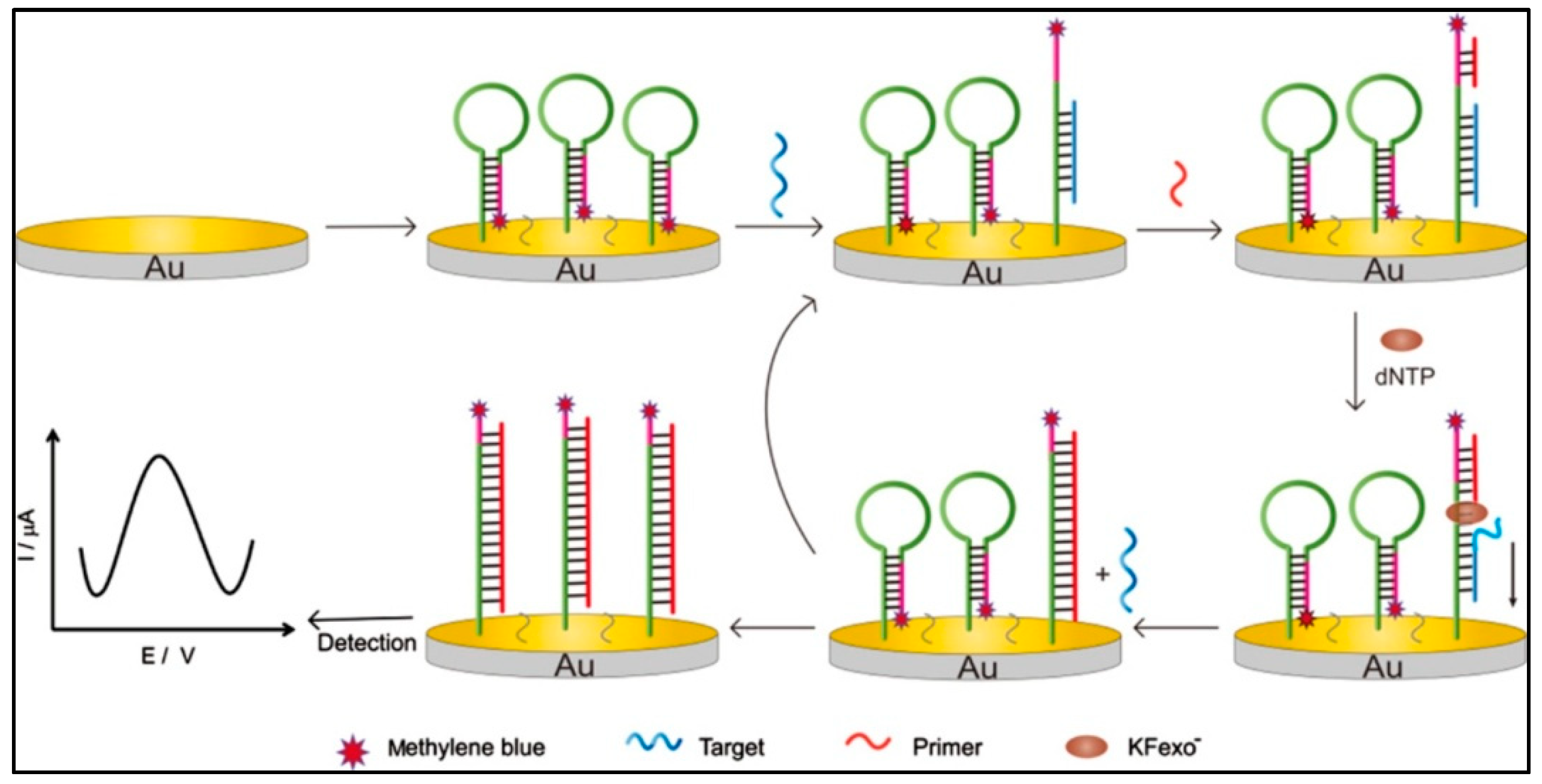
3.2.1. Target Cycle Amplification Technique
Exonuclease III-Assisted Target Cycle Amplification
Circular Strand-Replacement Polymerization
Catalyzed Hairpin Assembly
3.2.2. Hybridization Chain Reaction
3.2.3. DNA Isothermal Amplification Technology
Rolling Circle Amplification
Loop-Mediated Isothermal Amplification
Strand Displacement Amplification
4. Summary and Conclusions
5. Future Perspectives
Acknowledgments
Conflicts of Interest
References
- Singh, A.; Poshtiban, S.; Evoy, S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Agarwal, M.; Goswam, M.; Sharma, A.; Roy, S.; Rai, R.; Murugan, M. Biosensors: Tool for food borne pathogen detection. Vet. World 2013, 6, 968–973. [Google Scholar] [CrossRef]
- Wei, C.J.; Zhong, J.L.; Hu, T.; Zhao, X.H. Simultaneous detection of Escherichia coli O157:H7, Staphylococcus aureus and Salmonella by multiplex PCR in milk. 3 Biotech 2018, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Mani-López, E.; García, H.S.; López-Malo, A. Organic acids as antimicrobials to control Salmonella in meat and poultry products. Food Res. Int. 2012, 45, 713–721. [Google Scholar] [CrossRef]
- Zhou, K.; Zhong, K.C.; Long, C.; Han, X.F.; Liu, S.L. Development and Validation of a Predictive Model for the Growth of Salmonella enterica in Chicken Meat. J. Food Saf. 2014, 34, 326–332. [Google Scholar] [CrossRef]
- Georgescu, N.; Apostol, L.; Gherendi, F. Inactivation of Salmonella enterica serovar Typhimurium on egg surface, by direct and indirect treatments with cold atmospheric plasma. Food Control 2017, 76, 52–61. [Google Scholar] [CrossRef]
- Yu, Q.; Zhai, L.G.; Bei, X.M.; Liu, Z.X.; Zhang, C.; Tao, T.T.; Li, J.J.; Lv, F.X.; Zhao, H.Z. Survey of five food-borne pathogens in commercial cold food dishes and their detection by multiplex PCR. Food Control 2015, 59, 862–869. [Google Scholar] [CrossRef]
- Bardasi, L.; Taddei, R.; Nocera, L.; Ricchi, M.; Merialdi, G. Shiga Toxin-Producing Escherichia coli in Meat and Vegetable Products in Emilia Romagna Region, Years 2012–2013. Ital. J. Food Saf. 2015, 4, 4511. [Google Scholar] [CrossRef] [PubMed]
- Sofos, J.N.; Geornaras, I. Overview of current meat hygiene and safety risks and summary of recent studies on biofilms, and control of Escherichia coli O157:H7 in nonintact, and Listeria monocytogenes in ready-to-eat, meat products. Meat Sci. 2010, 86, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Abadias, M.; Alegre, I.; Oliveira, M.; Altisent, R.; Viñas, I. Growth potential of Escherichia coli O157:H7 on fresh-cut fruits (melon and pineapple) and vegetables (carrot and escarole) stored under different conditions. Food Control 2012, 27, 37–44. [Google Scholar] [CrossRef]
- Gullianklanian, M.; Sánchezsolis, J.M. Growth kinetics of Escherichia coli O157:H7 on the epicarp of fresh vegetables and fruits. Braz. J. Microbiol. 2018, 49, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Baloch, A.B.; Yang, H.; Feng, Y.; Xi, M.; Wu, Q.; Tang, J.; He, X.; Xiao, Y.; Xia, X. Presence and Antimicrobial Resistance of Escherichia coli in Ready-to-Eat Foods in Shaanxi, China. J. Food Prot. 2017, 80, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wu, Q.; Zhang, J.; Zhu, X. Occurrence and Characterization of Enteropathogenic Escherichia coli (EPEC) in Retail Ready-to-Eat Foods in China. Foodborne Pathog. Dis. 2015, 13, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Halabi, M.; Orth, D.; Grif, K.; Wiesholzer-pittl, M.; Kainz, M.; Schöberl, J.; Dierich, M.P.; Allerberger, F.; Würzner, R. Prevalence of Shiga toxin-, intimin- and haemolysin genes in Escherichia coli isolates from drinking water supplies in a rural area of Austria. Int. J. Hyg. Environ. Health 2008, 211, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Alcaine, S.; Jiang, Z.W.; M Rotello, V.; R Nugen, S. Detection of Escherichia coli in Drinking Water Using T7 Bacteriophage-Conjugated Magnetic Probe. Anal. Chem. 2015, 87, 8977–8984. [Google Scholar] [CrossRef] [PubMed]
- Perera, M.N.; Abuladze, T.; Li, M.R.; Woolston, J.; Sulakvelidze, A. Bacteriophage cocktail significantly reduces or eliminates Listeria monocytogenes contamination on lettuce, apples, cheese, smoked salmon and frozen foods. Food Microbiol. 2015, 52, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Mclennon, J.; Borza, A.; Eisebraun, M.; Garduño, R.A. Improved Recovery of Stressed Listeria monocytogenes from Frozen Foods. Food Anal. Methods 2018, 11, 403–414. [Google Scholar] [CrossRef]
- Mengesha, D.; Zewde, B.M.; Toquin, M.T.; Kleer, J.; Hildebrant, G.; Gebreves, W.A. Occurrence and distribution of Listeria monocytogenes and other Listeria species in ready-to-eat and raw meat products. Berliner Und Münchener Tierärztliche Wochenschrift 2009, 122, 20–24. [Google Scholar] [PubMed]
- Rodriguez-Lazaro, D.; Gonzalez-García, P.; Valero, A.; Hernandez, M. Application of the SureTect Detection Methods for Listeria monocytogenes and Listeria spp. in Meat, Dairy, Fish, and Vegetable Products. Food Anal. Methods 2015, 8, 1–6. [Google Scholar] [CrossRef]
- Nastou, A.; Jonathan, R.; Smirniotis, P.; Makri, L.; Kontominas, M.; Likotrafiti, E. Efficacy of household washing treatments for the control of Listeria monocytogenes on salad vegetables. Int. J. Food Microbiol. 2012, 159, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Muhammed, W.; Muleta, D.; Deneke, Y.; Gashaw, A.; Bitew, M. Studies on occurence of Listeria monocytogenes and other species in milk and milk products in retail market of Jimma town, Ethiopia. Asian J. Dairy Food Res. 2013, 32, 35–39. [Google Scholar]
- Salazar, J.K.; Bathija, V.M.; Carstens, C.K.; Narula, S.S.; Shazer, A.; Stewart, D.; Tortotello, M.L. Listeria monocytogenes Growth Kinetics in Milkshakes Made from Naturally and Artificially Contaminated Ice Cream. Front. Microbiol. 2018, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Wernars, K.; Heuvelman, C.J.; Chakraborty, T.; Notermans, S.H.W. Use of the polymerase chain reaction for direct detection of Listeria monocytogenes in soft cheese. J. Appl. Microbiol. 2010, 70, 121–126. [Google Scholar]
- Zhang, Z.H.; Liu, W.T.; Xu, H.Y.; Aguilar, Z.P.; Shah, N.P.; Wei, H. Propidium monoazide combined with real-time PCR for selective detection of viable Staphylococcus aureus in milk powder and meat products. J. Dairy Sci. 2015, 98, 1625–1633. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.I.; Joo, I.S.; Choi, J.H.; Jung, K.H.; Choi, E.J.; Son, N.R.; Han, M.K.; Jeong, S.J.; Lee, S.H.; Hwang, I.G. Distribution of Methicillin-resistant Staphylococcus aureus (MRSA) in RAW Meat and Fish Samples in Korea. Food Sci. Biotechnol. 2014, 23, 999–1003. [Google Scholar] [CrossRef]
- Can, H.Y.; Elmalı, M.; Karagöz, A. Molecular Typing and Antimicrobial Susceptibility of Staphylococcus aureus Strains Isolated from Raw Milk, Cheese, Minced Meat, and Chicken Meat Samples. Korean J. Food Sci. Anim. Resour. 2017, 37, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.P.; Li, M.Y.; Zhao, G.M.; Cui, W.M.; Gao, X.P.; Liu, Y.X.; Zhao, L.J.; Wang, Y.F. Thermal Inactivation Properties of Shigella in Smoked Cooked Ham. Food Sci. 2017, 38, 40–45. [Google Scholar]
- Shin, W.S.; Hong, W.S.; Lee, K.E. Assessment of Microbiological Quality for Raw Materials and Cooked Foods in Elementary School Food Establishment. J. Korean Soc. Food Sci. Nutr. 2008, 37, 379–389. [Google Scholar] [CrossRef]
- Shahin, K.; Bouzari, M. Bacteriophage application for biocontrolling Shigella flexneri in contaminated foods. J. Food Sci. Technol. 2018, 55, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Sonbol, H.; Joseph, S.; McAuley, C.M.; Craven, H.M.; Forsythe, S.J. Multilocus sequence typing of Cronobacter spp. from powdered infant formula and milk powder production factories. Int. Dairy J. 2013, 30, 1–7. [Google Scholar] [CrossRef]
- Pinapérez, M.C.; Rodrigo, D.; Martínez, A. Non-Thermal Inactivation of Cronobacter Sakazakii in Infant Formula Milk: A Review. Crit. Rev. Food Sci. Nutr. 2015, 56, 1620–1629. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.Z.; Gao, J.X.; Liu, Y.; Chen, H.Y. Isolation of Cronobacter spp. isolates from infant formulas and their survival in the production process of infant formula. Czech J. Food Sci. 2011, 29, 391–399. [Google Scholar] [CrossRef]
- Forsythe, S.J. Enterobacter sakazakii and other bacteria in powdered infant milk formula. Matern. Child Nutr. 2010, 1, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Azwai, S.M.; Alfallani, E.A.; Abolqhait, S.K.; Garbaj, A.M.; Naas, H.T.; Moawad, A.A.; Gammoudi, F.T.; Rayes, H.M.; Barbieri, I.; Eldaghayes, I.M. Isolation and molecular identification of Vibrio spp. by sequencing of 16S rDNA from seafood, meat and meat products in Libya. Open Vet. J. 2016, 6, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Dileep, V.; Sanath, K.; Yadhu, K.; Mitsuaki, N.; Indrani, K.; Iddya, K. Application of polymerase chain reaction for detection of Vibrio parahaemolyticus associated with tropical seafoods and coastal environment. Lett. Appl. Microbiol. 2010, 36, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.W.; Lee, S.H.; Hwang, I.G.; Yoon, K.S. Effect of Temperature on Growth of Vibrio paraphemolyticus and Vibrio vulnificus in Flounder, Salmon Sashimi and Oyster Meat. Int. J. Environ. Res. Public Health 2012, 9, 4662–4675. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, S.Y.; Yu, J.Y.; Zhang, H.; Yuan, Z.Q.; Sun, Y.S.; Zhang, L.; Zhu, Y.F.; Song, H.B. An outbreak of Proteus mirabilis food poisoning associated with eating stewed pork balls in brown sauce, Beijing. Food Control 2010, 21, 302–305. [Google Scholar] [CrossRef]
- Adedeji, B.S.; Ezeokoli, O.T.; Ezekiel, C.N.; Obadina, A.O.; Somorin, Y.M.; Sulyok, M.; Adeleke, R.A.; Warth, B.; Nwangburuka, C.C.; Omemu, A.M.; et al. Bacterial species and mycotoxin contamination associated with locust bean, melon and their fermented products in south-western Nigeria. Int. J. Food Microbiol. 2017, 258, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Tompkin, R.B.; Christiansen, L.N.; Shaparis, A.B. Enhancing nitrite inhibition of Clostridium botulinum with isoascorbate in perishable canned cured meat. Appl. Environ. Microbiol. 1978, 35, 59. [Google Scholar] [PubMed]
- Roberts, T.A.; Gibson, A.M.; Robinson, A. Factors controlling the growth of Clostridium botulinum types A and B in pasteurized, cured meats. Int. J. Food Sci. Technol. 2010, 17, 307–326. [Google Scholar] [CrossRef]
- Hospital, X.F.; Hierro, E.; Stringer, S.; Fernández, M. A study on the toxigenesis by Clostridium botulinum in nitrate and nitrite-reduced dry fermented sausages. Int. J. Food Microbiol. 2015, 218, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Delbrassinne, L.; Andjelkovic, M.; Dierick, K.; Denayer, S.; Mahillon, J.; Loco, J.V. Prevalence and levels of Bacillus cereus emetic toxin in rice dishes randomly collected from restaurants and comparison with the levels measured in a recent foodborne outbreak. Foodborne Pathog. Dis. 2012, 9, 809–814. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, D.; Fortunato, F.; Tafuri, S.; Cozza, V.; Chironna, M.; Germinario, C.; Pedalino, B.; Prato, R. Lessons learnt from a birthday party: A Bacillus cereus outbreak, Bari, Italy, January 2012. Ann. Ist. Super. Sanita 2013, 49, 391–394. [Google Scholar] [PubMed]
- Delbrassinne, L.; Botteldoorn, N.; Andjelkovic, M.; Dierick, K.; Denayer, S. An emetic Bacillus cereus outbreak in a kindergarten: Detection and quantification of critical levels of cereulide toxin. Foodborne Pathog. Dis. 2015, 12, 84. [Google Scholar] [CrossRef] [PubMed]
- Sallam, K.I. Prevalence of Campylobacter in chicken and chicken by-products retailed in Sapporo area, Hokkaido, Japan. Food Control 2007, 18, 1113–1120. [Google Scholar] [CrossRef]
- Whyte, R.; Hudson, J.A.; Graham, C. Campylobacter in chicken livers and their destruction by pan frying. Lett. Appl. Microbiol. 2010, 43, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Muniandy, S.; The, S.J.; Thong, K.L.; Thiha, A.; Dinshaw, I.J.; Lai, C.W.; Ibrahim, F.; Leo, B.F. Carbon Nanomaterial-Based Electrochemical Biosensors for Foodborne Bacterial Detection. Crit. Rev. Anal. Chem. 2019, 49, 510–533. [Google Scholar] [CrossRef] [PubMed]
- Mullis, K.B.; Ferré, F.; Gibbs, R.A. The Polymerase Chain Reaction. Infect. Dis. News. 1994, 8, 36–39. [Google Scholar]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, B.; Roberts, S.; Grimwade, B.; Shao, W.P.; Wang, M.J.; Fu, Q.; Shu, Q.P.; Laroche, I.; Zhou, Z.M.; Tchernev, V.T.; et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat. Biotechnol. 2002, 20, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.F.D.; Magalhãesb, J.M.C.S.; Freirec, C.; Delerue-Matosa, C. Electrochemical biosensors for Salmonella: State of the art and challenges in food safety assessment. Biosens. Bioelectron. 2018, 99, 667–682. [Google Scholar] [CrossRef] [PubMed]
- Reta, N.; Saint, C.P.; Michelmore, A.; Prieto-Simon, B.; Voelcker, N.H. Nanostructured Electrochemical Biosensors for Label-Free Detection of Water- and Food-Borne Pathogens. ACS Appl. Mater. Interfaces 2018, 10, 6055–6072. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Sindhu, A.; Dilbaghi, N.; Chaudhury, A. Biosensors as innovative tools for the detection of food borne pathogens. Biosens. Bioelectron. 2011, 28, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rafique, B.; Iqbal, M.; Mehmood, T.; Shaheen, M.A. Electrochemical DNA biosensors: A review. Sens. Rev. 2019, 39, 34–50. [Google Scholar] [CrossRef]
- Liu, S.; Zheng, Z.; Li, X. Advances in pesticide biosensors: Current status, challenges, and future perspectives. Anal. Bioanal. Chem. 2013, 405, 63–90. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.D.; Martin, L.J.; Simpson, R.B.; Wallis, D.E. Comparison of polymerase chain reaction and microbiological culture for detection of salmonellae in equine feces and environmental samples. Am. J. Vet. Res. 1996, 57, 780. [Google Scholar] [PubMed]
- Ngwa, G.A.; Schop, R.; Weir, S.; León-Velarde, C.G.; Odumeru, J.A. Detection and enumeration of E. coli O157:H7 in water samples by culture and molecular methods. J. Microbiol. Methods 2013, 92, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Poli, M.A.; Rivera, V.R.; Neal, D. Sensitive and specific colorimetric ELISAs for Staphylococcus aureus enterotoxins A and B in urine and buffer. Toxicon 2002, 40, 1723–1726. [Google Scholar] [CrossRef]
- Chunglok, W.; Wuragil, D.K.; Oaewc, S.; Somasundrumc, M.; Surareungchai, W. Immunoassay based on carbon nanotubes-enhanced ELISA for Salmonella enterica serovar Typhimurium. Biosens. Bioelectron. 2011, 26, 3584–3589. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Chen, X.Z.; Li, K.S.; Du, H.F.; Chen, Y.T.; Lian, X.W.; Lu, X.P.; Yuan, M.; Ye, W.H. Extraction and identification of E. coli O157∶H7 lipopolysaccharide and development of an indirect ELISA. Chin. J. Zoonoses 2011, 27, 637–640. [Google Scholar]
- Zhu, F.; Rogelj, S.; Kieft, T.L. Rapid detection of Escherichia coli O157:H7 by immunomagnetic separation and real-time PCR. Int. J. Food Microbiol. 2011, 99, 47–57. [Google Scholar]
- Chen, Z.Z.; Cai, L.; Chen, M.Y.; Lin, Y.; Pang, D.W.; Tang, H.W. Indirect immunofluorescence detection of E. coli O157:H7 with fluorescent silica nanoparticles. Biosens. Bioelectron. 2015, 66, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Lang, J.; Shen, Z.Q.; Chen, Z.L. A Rapid Subtractive Immunization Method to Prepare Discriminatory Monoclonal Antibodies for Food E. coli O157:H7 Contamination. PLoS ONE 2012, 7, e31352. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, H.R.; Najafi, A.; Ahmadi, A. Rapid, specific and concurrent detection of Listeria, Salmonella and Escherichia coli pathogens by multiplex PCR in Iranian food. Afr. J. Microbiol. Res. 2010, 4, 2503–2507. [Google Scholar]
- Malorny, B.; Bunge, C.; Helmuth, R. A real-time PCR for the detection of Salmonella Enteritidis in poultry meat and consumption eggs. J. Microbiol. Methods 2007, 70, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Rhim, S.R.; Kim, K.T.; Paik, H.D.; Lee, J.Y. Simultaneous Detection of Listeria monocytogenes, Escherichia coli O157:H7, Bacillus cereus, Salmonella spp., and Staphylococcus aureus in Low-fatted Milk by Multiplex PCR. Korean J. Food Sci. Anim. Resour. 2014, 34, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Arunrut, N.; Kiatpathomchai, W.; Ananchaipattana, C. Multiplex PCR assay and lyophilization for detection of Salmonella spp., Staphylococcus aureus and Bacillus cereus in pork products. Food Sci. Biotechnol. 2018, 27, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Kwon, K.Y.; Oh, S.K.; Chang, H. A multiplex PCR assay for simultaneous detection of Escherichia coli O157:H7, Bacillus cereus, Vibrio parahaemolyticus, Salmonella spp., Listeria monocytogenes, and Staphylococcus aureus in Korean ready-to-eat food. Foodborne Pathog. Dis. 2014, 11, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yan, L.N.; Wu, X.; Li, F.; Wang, D.; Xu, H.Y. Multiplex PCR coupled with propidium monoazide for the detection of viable Cronobacter sakazakii, Bacillus cereus, and Salmonella spp. in milk and milk products. J. Dairy Sci. 2017, 100, 7874. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.J.; Ma, L.M.; Zheng, S.M.; He, X.H.; Hammack, T.S.; Brown, E.W.; Zhang, G.D. Development of a novel loop-mediated isothermal amplification (LAMP) assay for the detection of Salmonella ser. Enteritidis from egg products. Food Control 2018, 88, 190–197. [Google Scholar] [CrossRef]
- Kordas, A.; Papadakis, G.; Milioni, D.; Champ, J.; Descroix, S.; Gizeli, E. Rapid Salmonella detection using an acoustic wave device combined with the RCA isothermal DNA amplification method. Sens. Bio-Sens. Res. 2016, 11, 121–127. [Google Scholar] [CrossRef]
- Ãsterberg, F.W.; Rizzi, G.; Donolato, M.; Bejhed, R.S.; Mezger, A.; Strömberg, M.; Nilsson, M.; Strømme, M.; Svedlindh, P.; Hansen, M.; et al. On-chip detection of rolling circle amplified DNA molecules from Bacillus globigii spores and Vibrio cholerae. Small 2014, 10, 2877–2882. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, Q.; Zhang, Y.Z.; Meng, Z.X.; Ma, X.Y.; Zhang, W. Saltatory Rolling Circle Amplification (SRCA): A Novel Nucleic Acid Isothermal Amplification Technique Applied for Rapid Detection of Shigella spp. in Vegetable Salad. Food Anal. Methods 2018, 11, 504–513. [Google Scholar] [CrossRef]
- Shukla, S.; Haldoraia, Y.; Bajpaia, V.K.; Rengarajc, A.; Hwangc, S.K.; Song, X.J.; Kimd, M.; Huhc, Y.S.; Hana, Y.K. Electrochemical coupled immunosensing platform based on graphene oxide/gold nanocomposite for sensitive detection of Cronobacter sakazakii in powdered infant formula. Biosens. Bioelectron. 2018, 109, 139. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.R.; Reddy, B.M.; Sunki, G.R.; Pulusani, S.R. Influence of antimicrobial compound(s) extracted from milk fermented by Streptococcus thermophilus on keeping quality of meat and milk. J. Food Qual. 2010, 4, 247–258. [Google Scholar] [CrossRef]
- Izadi, Z.; Sheikh-Zeinoddin, M.; Ensafi, A.A.; Soleimanian-Zad, S. Fabrication of an electrochemical DNA-based biosensor for Bacillus cereus detection in milk and infant formula. Biosens. Bioelectron. 2016, 80, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Eivazzadeh-Keihan, R.; Pashazadeh-Panahi, P.; Baradaran, B.; Guardia, M.D.L.; Hejazi, M.; Sohrabi, H.; Mokhtarzadeh, A.; Maleki, A. Recent progress in optical and electrochemical biosensors for sensing of Clostridium botulinum neurotoxin. TrAC Trends Anal. Chem. 2018, 103, 184–197. [Google Scholar] [CrossRef]
- Holowko, M.B.; Poh, C.L. Designing and Assembling Plasmids for the Construction of Escherichia coli Biosensor for Vibrio cholerae Detection. Methods Mol. Biol. 2018, 1772, 445. [Google Scholar] [PubMed]
- Abdalhai, M.H.; Fernandes, A.M.; Xia, X.; Musa, A.; Ji, J.; Sun, X. Electrochemical Genosensor to Detect Pathogenic Bacteria (Escherichia coli O157:H7) As Applied in Real Food Samples (Fresh Beef) To Improve Food Safety and Quality Control. J. Agric. Food Chem. 2015, 63, 5017–5025. [Google Scholar] [CrossRef] [PubMed]
- Nazari-Vanani, R.; Sattarahmady, N.; Yadegari, H.; Heli, H. A novel and ultrasensitive electrochemical DNA biosensor based on an ice crystals-like gold nanostructure for the detection of Enterococcus faecalis gene sequence. Colloids Surf. B Biointerfaces 2018, 166, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Yan, Y.R.; Lei, P.H.; Shen, B.; Cheng, W.; Ju, H.X.; Ding, S.J. A novel electrochemical sensing strategy for rapid and ultrasensitive detection of Salmonella by rolling circle amplification and DNA-AuNPs probe. Anal. Chim. Acta 2014, 846, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Benvidi, A.; Tezerjani, M.D.; Jahanbani, S.; Ardakani, M.M.; Moshtaghioun, S.M. Comparison of impedimetric detection of DNA hybridization on the various biosensors based on modified glassy carbon electrodes with PANHS and nanomaterials of RGO and MWCNTs. Talanta 2016, 147, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Low, K.F.; Chuenrangsikul, K.; Rijiravanich, P.; Suraneungchai, W.; Chan, Y.Y. Electrochemical genosensor for specific detection of the food-borne pathogen, Vibrio cholerae. World J. Microbiol. Biotechnol. 2012, 28, 1699–1706. [Google Scholar] [CrossRef] [PubMed]
- Hassani, S.; Akmalc, M.R.; Salek-Maghsoudib, A.; Rahmania, S.; Ganjalic, M.R.; Norouzic, P.; Abdollahia, M. Novel label-free electrochemical aptasensor for determination of Diazinon using gold nanoparticles-modified screen-printed gold electrode. Biosens. Bioelectron. 2018, 120, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Qi, X.W.; Zhang, Y.Y.; Yang, H.R.; Gao, H.W.; Chen, Y.; Sun, Z.F. Electrochemical DNA biosensor for the detection of Listeria monocytogenes with dendritic nanogold and electrochemical reduced graphene modified carbon ionic liquid electrode. Electrochim. Acta 2012, 85, 145–151. [Google Scholar] [CrossRef]
- Gaudin, V. Advances in biosensor development for the screening of antibiotic residues in food products of animal origin—A comprehensive review. Biosens. Bioelectron. 2017, 90, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434. [Google Scholar] [CrossRef] [PubMed]
- Lönne, M.; Zhu, G.H.; Stahl, F.; Walter, J.G. Aptamer-Modified Nanoparticles as Biosensors. Adv. Biochem. Eng. Biotechnol. 2014, 140, 121–154. [Google Scholar] [PubMed]
- Bini, A.; Centi, S.; Tombelli, S.; Minunni, M. Development of an optical RNA-based aptasensor for C-reactive protein. Anal. Bioanal. Chem. 2008, 390, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Jiang, Y.H.; Jia, F.; Yu, Y.; Chen, J.; Wang, Z.P. An aptamer-based electrochemical biosensor for the detection of Salmonella. J. Microbiol Methods 2014, 98, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ou, L.J.; Chu, X.; Shen, G.L. An electrochemical aptasensor based on gold nanoparticles dotted graphene modified glassy carbon electrode for label-free detection of bisphenol A in milk samples. Food Chem. 2014, 162, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Fang, Z.Y.; Zhao, S.M.; Lu, X.W. A simple aptamer biosensor for Salmonellae enteritidis based on fluorescence-switch signaling graphene oxide. RSC Adv. 2014, 4, 22009–22012. [Google Scholar] [CrossRef]
- Lee, H.; Kim, B.C.; Kim, K.W.; Kim, Y.K.; Kim, J.B.; Oh, M.K. A sensitive method to detect Escherichia coli based on immunomagnetic separation and real-time PCR amplification of aptamers. Biosens. Bioelectron. 2009, 24, 3550–3555. [Google Scholar] [CrossRef] [PubMed]
- Ozalp, V.C.; Bayramoglu, G.; Kavruk, M.; Keskin, B.B.; Oktem, H.A.; Arica, M.Y. Pathogen detection by core–shell type aptamer-magnetic preconcentration coupled to real-time PCR. Anal. Biochem. 2014, 447, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.B.; Chai, Y.Q.; Yuan, Y.L.; Bai, L.J.; Yuan, R. Development of an electrochemical method for Ochratoxin A detection based on aptamer and loop-mediated isothermal amplification. Biosens. Bioelectron. 2014, 55, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Y.; Chang, Y.W.; Tai, Y.T.; Yang, Y.S. High sensitive, colorimetric, isothermal nucleic acids amplification: A versatile platform for protein biosensors. In Proceedings of the International Conference on Sensing Technology, Tainan, Taiwan, 30 November–3 December 2008. [Google Scholar]
- Yang, L.L.; Zhang, Y.; Li, R.B.; Lin, C.Y.; Guo, L.H.; Qiu, B.; Lin, Z.Y.; Chen, G.N. Electrochemiluminescence biosensor for ultrasensitive determination of ochratoxin A in corn samples based on aptamer and hyperbranched rolling circle amplification. Biosens. Bioelectron. 2015, 70, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ou, L.J.; Chu, X.; Shen, G.L. Aptamer-Based Rolling Circle Amplification: A Platform for Electrochemical Detection of Protein. Anal. Chem. 2007, 79, 7492–7500. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.M.; Bai, W.H.; Zhu, C.; Huang, Y.F.; Yan, Y.J.; Chen, A.L. Design of nuclease-based target recycling signal amplification in aptasensors. Biosens. Bioelectron. 2016, 77, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Yuan, R.; Yi, L.; Yang, J.L.; Zhang, H.W.; Lia, L.X.; Nian, W.Q.; Yi, G. Target-induced aptamer displacement on gold nanoparticles and rolling circle amplification for ultrasensitive live Salmonella typhimurium electrochemical biosensing. J. Electroanal. Chem. 2018, 826, 174–180. [Google Scholar] [CrossRef]
- Rashid, J.I.A.; Yusof, N.A. The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: A review. Sens. Bio-Sens. Res. 2017, 16, 19–31. [Google Scholar] [CrossRef]
- Velusamy, V.; Arshak, K.; Yang, C.F.; Yu, L.; Korostynska, O.; Adley, C. Comparison between DNA Immobilization Techniques on a Redox Polymer Matrix. Am. J. Anal. Chem. 2011, 2, 392–400. [Google Scholar] [CrossRef]
- Amouzadeh Tabrizi, M.; Shamsipur, M. A label-free electrochemical DNA biosensor based on covalent immobilization of salmonella DNA sequences on the nanoporous glassy carbon electrode. Biosens. Bioelectron. 2015, 69, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Sheikhzadeh, E.; Chamsaz, M.; Turner, A.P.F.; Jager, E.W.H.; Beni, V. Label-free impedimetric biosensor for Salmonella Typhimurium detection based on poly [pyrrole-co-3-carboxyl-pyrrole] copolymer supported aptamer. Biosens. Bioelectron. 2016, 80, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Arora, K.; Prabhakar, N.; Chand, S.; Malhotra, B.D. Immobilization of single stranded DNA probe onto polypyrrole-polyvinyl sulfonate for application to DNA hybridization biosensor. Sens. Actuators B Chem. 2007, 126, 655–663. [Google Scholar] [CrossRef]
- Arora, K.; Prabhakar, N.; Chand, S.; Malhotra, B.D. Escherichia coli Genosensor Based on Polyaniline. Anal. Chem. 2007, 79, 6152–6158. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Duan, N.; Wu, S.J.; Ma, X.Y.; Xia, Y.; Wang, Z.P.; Wei, X.L. Impedimetric aptasensor for Staphylococcus aureus based on nanocomposite prepared from reduced graphene oxide and gold nanoparticles. Microchim. Acta 2014, 181, 967–974. [Google Scholar] [CrossRef]
- Reich, P.; Stoltenburg, R.; Strehlitz, B.; Frense, D.; Beckmann, D. Development of an Impedimetric Aptasensor for the Detection of Staphylococcus aureus. Int. J. Mol. Sci. 2017, 18, 2484. [Google Scholar] [CrossRef] [PubMed]
- Andrade, C.; Nascimento, J.M.; Oliveira, I.S.; De Oliveira, C.V.J. Nanostructured sensor based on carbon nanotubes and clavanin A for bacterial detection. Colloids Surf. B Biointerfaces 2015, 135, 833–839. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, J.L.; Oliveira, M.D.L.; Oliveira, I.S.; Frias, I.A.M.; Franco, O.L.; Andrade, C.A.S. A simple nanostructured biosensor based on clavanin A antimicrobial peptide for gram-negative bacteria detection. Biochem. Eng. J. 2017, 124, 108–114. [Google Scholar] [CrossRef]
- Singh, A.; Sinsinbar, G.; Choudhary, M.; Kumar, V.; Pasricha, R.; Verma, H.N.; Singh, S.P.; Arora, K. Graphene oxide-chitosan nanocomposite based electrochemical DNA biosensor for detection of typhoid. Sens. Actuators B Chem. 2013, 185, 675–684. [Google Scholar] [CrossRef]
- Tiwari, I.; Singh, M.; Pandey, C.M.; Sumana, G. Electrochemical genosensor based on graphene oxide modified iron oxide–chitosan hybrid nanocomposite for pathogen detection. Sens. Actuators B Chem. 2015, 206, 276–283. [Google Scholar] [CrossRef]
- Mazzaracchio, V.; Neagu, D.; Porchetta, A.; Marcoccio, E.; Pomponi, A.; Faggioni, G.; D’Amore, N.; Notargiacomo, A.; Pea, M.; Moscone, D.; et al. A label-free impedimetric aptasensor for the detection of Bacillus anthracis spore simulant. Biosens. Bioelectron. 2019, 126, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Ligaj, M.; Tichoniuk, M.; Gwiazdowska, D.; Filipiak, M. Electrochemical DNA biosensor for the detection of pathogenic bacteria Aeromonas hydrophila. Electrochim. Acta 2014, 128, 67–74. [Google Scholar] [CrossRef]
- Vanegas, D.C.; Rong, Y.; Schwalb, N.; Hills, K.D.; Gomes, C.; McLamore, E.S. Rapid detection of listeria spp. using an internalin A aptasensor based on carbon-metal nanohybrid structures. In Proceedings of the SPIE Sensing Technology + Applications, Baltimore, MD, USA, 13 May 2015; Volume 9487, p. 948708. [Google Scholar]
- Singh, A.; Choudhary, M.; Singh, M.P.; Verma, H.N.; Singh, S.P.; Arora, K. DNA Functionalized Direct Electro-deposited Gold nanoaggregates for Efficient Detection of Salmonella typhi. Bioelectrochemistry 2015, 105, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.H.; Tang, H.; Cheng, W.; Yan, Y.; Zhang, D.C.; Ju, H.X.; Ding, S.J. A sensitive electrochemical DNA biosensor for specific detection of Enterobacteriaceae bacteria by Exonuclease III-assisted signal amplification. Biosens. Bioelectron. 2013, 48, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Fu, H.; He, Y.; Zhai, Q.; Guo, J.; Qing, K.Q.; Yi, G. Electrochemical Aptasensor for Rapid and Sensitive Determination of Salmonella Based on Target-Induced Strand Displacement and Gold Nanoparticle Amplification. Anal. Lett. 2016, 49, 2405–2417. [Google Scholar] [CrossRef]
- Hu, Y.H.; Xu, X.Q.; Liu, Q.H.; Wang, L.; Lin, Z.Y.; Chen, G.N. Ultrasensitive electrochemical biosensor for detection of DNA from Bacillus subtilis by coupling target-induced strand displacement and nicking endonuclease signal amplification. Anal. Chem. 2014, 86, 8785–8790. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Duan, N.; Wu, S.J.; Dai, R.T.; Wang, Z.P.; Li, X.M. Impedimetric Salmonella aptasensor using a glassy carbon electrode modified with an electrodeposited composite consisting of reduced graphene oxide and carbon nanotubes. Microchim. Acta 2016, 183, 337–344. [Google Scholar] [CrossRef]
- Hasan, M.R.; Pulingam, T.; Appaturi, J.N.; Zifruddin, A.N.; Teh, S.J.; Lim, T.W.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Carbon nanotube-based aptasensor for sensitive electrochemical detection of whole-cell Salmonella. Anal. Biochem. 2018, 554, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Qin, P.; Gao, H.W.; Li, G.C.; Jiao, K. Electrochemical DNA biosensor based on chitosan/nano-V2O5/MWCNTs composite film modified carbon ionic liquid electrode and its application to the LAMP product of Yersinia enterocolitica gene sequence. Biosens. Bioelectron. 2010, 25, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Pei, Q.Q.; Wang, Y.; Liu, S.; Qin, Y.F.; Leng, X.Q.; Cui, X.J.; Huang, J.D. Exonuclease III-aided autonomous cascade signal amplification: A facile and universal DNA biosensing platform for ultrasensitive electrochemical detection of S. typhimurium. New J. Chem. 2017, 41, 7613–7620. [Google Scholar] [CrossRef]
- Bai, Y.H.; Li, J.Y.; Xu, J.J.; Chen, H.Y. Ultrasensitive electrochemical detection of DNA hybridization using Au/Fe3O4 magnetic composites combined with silver enhancement. Analyst 2010, 135, 1672–1679. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.S.; Ju, H.X.; Chen, H.Y. Voltammetric Behavior and Detection of DNA at Electrochemically Pretreated Glassy Carbon Electrode. Electroanalysis 2015, 13, 1105–1109. [Google Scholar] [CrossRef]
- Thorp and, H.H. Cutting out the middleman: DNA biosensors based on electrochemical oxidation. Trends Biotechnol. 1998, 16, 117–121. [Google Scholar] [CrossRef]
- Armistead, P.M.; Thorp, H.H. Electrochemical Detection of Gene Expression in Tumor Samples: Overexpression of Rak Nuclear Tyrosine Kinase. Bioconjug. Chem. 2002, 13, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Azimzadeh, M.; Rahaie, M.; Nasirizadeh, N.; Ashtari, K.; Naderi-Manesh, H. An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosens. Bioelectron. 2016, 77, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Bai, W.Q.; Dong, C.X.; Guo, R.; Liu, Z.H. An ultrasensitive electrochemical DNA biosensor based on graphene/Au nanorod/polythionine for human papillomavirus DNA detection. Biosens. Bioelectron. 2015, 68, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.K.; Yan, W.W.; Liu, Y.Q.; He, S.D.; Cao, X.D.; Xu, X.; Zheng, H.S.; Gunasekaran, S. Electrochemical detection of Salmonella using an invA genosensor on polypyrrole-reduced graphene oxide modified glassy carbon electrode and AuNPs-horseradish peroxidase-streptavidin as nanotag. Anal. Chim. Acta 2019, 1074, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Hajihosseini, S.; Nasirizadeh, N.; Hejazi, M.S.; Yaghmaei, P. A sensitive DNA biosensor fabricated from gold nanoparticles and graphene oxide on a glassy carbon electrode. Mater. Sci. Eng. C 2016, 61, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Gholivand, M.B.; Akbari, A. A sensitive electrochemical genosensor for highly specific detection of thalassemia gene. Biosens. Bioelectron. 2019, 129, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Shi, A.Q.; Wang, J.; Han, X.W.; Fang, X.; Zhang, Y.Z. A sensitive electrochemical DNA biosensor based on gold nanomaterial and graphene amplified signal. Sens. Actuators B Chem. 2014, 200, 206–212. [Google Scholar] [CrossRef]
- Muniandy, S.; Teh, S.J.; Appaturi, J.N.; Thong, K.L.; Lai, C.W.; Ibrahim, F.; Leo, B.F. A reduced graphene oxide-titanium dioxide nanocomposite based electrochemical aptasensor for rapid and sensitive detection of Salmonella enterica. Bioelectrochemistry 2019, 127, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Benvidi, A.; Firouzabadi, A.D.; Moshtaghiun, S.M.; Mazloum-Ardakani, M.; Tezerjani, M.D. Ultrasensitive DNA sensor based on gold nanoparticles/reduced graphene oxide/glassy carbon electrode. Anal. Biochem. 2015, 484, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Raston, N.H.A.; Gu, M.B. Aptamer-based nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. [Google Scholar] [PubMed]
- Wei, B.; Mao, K.; Liu, N.; Zhang, M.; Yang, Z.G. Graphene nanocomposites modified electrochemical aptamer sensor for rapid and highly sensitive detection of prostate specific antigen. Biosens. Bioelectron. 2018, 121, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Iijima, S. Helical microtubules of graphitic carbon. Nature 1991, 354, 56–58. [Google Scholar] [CrossRef]
- Lan, L.Y.; Yao, Y.; Ping, J.F.; Ying, Y.B. Recent advances in nanomaterial-based biosensors for antibiotics detection. Biosens. Bioelectron. 2017, 91, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Jafari, S.; Faridbod, F.; Norouzi, P.; Dezfuli, A.S.; Ajloo, D. Fatemeh Mohammadipanah, Mohammad Reza Ganjali. Detection of Aeromonas hydrophila DNA oligonucleotide Sequence using a Biosensor Design based on Ceria Nanoparticles Decorated Reduced Graphene Oxide and Fast Fourier Transform Square Wave Voltammetry. Anal. Chim. Acta 2015, 895, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; He, X.X.; Ji, J.; Jia, M.; Wang, Z.P.; Sun, X.L. A highly selective and sensitive electrochemical CS-MWCNTs/Au-NPs composite DNA biosensor for Staphylococcus aureus gene sequence detection. Talanta 2015, 141, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.K.; Liu, Y.Q.; He, S.D.; Xu, X.; Cao, X.D.; Ye, Y.W.; Zheng, H.S. Ultrasensitive electrochemical DNA sensor for virulence invA gene of Salmonella using silver nanoclusters as signal probe. Sens. Actuators B Chem. 2018, 272, 53–59. [Google Scholar] [CrossRef]
- Sun, W.; Wang, X.L.; Lu, Y.X.; Gong, S.X.; Qi, X.W.; Lei, B.X.; Sun, Z.F.; Li, G.J. Electrochemical deoxyribonucleic acid biosensor based on electrodeposited graphene and nickel oxide nanoparticle modified electrode for the detection of salmonella enteritidis gene sequence. Mater. Sci. Eng. C 2015, 49, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, J.; Fang, L.C.; Yu, K.K.; Huang, H.; Jiang, L.L.; Liang, W.B.; Zheng, J.S. A novel electrochemical DNA biosensor based on HRP-mimicking hemin/G-quadruplex wrapped GOx nanocomposites as tag for detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2015, 63, 1. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.W.; Gao, H.W.; Zhang, Y.Y.; Wang, X.Z.; Chen, Y.; Sun, W. Electrochemical DNA biosensor with chitosan-Co(3)O(4) nanorod-graphene composite for the sensitive detection of Staphylococcus aureus nuc gene sequence. Bioelectrochemistry 2012, 88, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, Z.; Li, Y.Y.; Xie, G.M. Amplified electrochemical detection of mecA gene in methicillin-resistant Staphylococcus aureus based on target recycling amplification and isothermal strand-displacement polymerization reaction. Sens. Actuators B Chem. 2015, 221, 148–154. [Google Scholar] [CrossRef]
- Gao, F.L.; Zhu, Z.; Lei, J.P.; Geng, Y.; Ju, H.X. Sub-femtomolar electrochemical detection of DNA using surface circular strand-replacement polymerization and gold nanoparticle catalyzed silver deposition for signal amplification. Biosens. Bioelectron. 2013, 39, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Tang, D.Q.; Du, L.L.; Zhang, Y.Z.; Zhang, L.X.; Gao, F.L. A novel signal-on electrochemical DNA sensor based on target catalyzed hairpin assembly strategy. Biosens. Bioelectron. 2015, 64, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.S.; Lima, A.S.; Battistel, D.; Daniele, S.; Bertotti, M. Fabrication and Use of Dual-function Iridium Oxide Coated Gold SECM Tips. An Application to pH Monitoring above a Copper Electrode Surface during Nitrate Reduction. Electroanalysis 2016, 28, 1441–1447. [Google Scholar] [CrossRef]
- Choi, H.M.T.; Beck, V.A.; Pierce, N.A. Next-Generation in Situ Hybridization Chain Reaction: Higher Gain, Lower Cost, Greater Durability. ACS Nano 2014, 8, 4284–4294. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Cui, X.; Deng, Y.; Tang, Z. Amplified detection of nucleic acid by G-quadruplex based hybridization chain reaction. Biosens. Bioelectron. 2012, 38, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Dirks, R.M.; Pierce, N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 2004, 101, 15275–15278. [Google Scholar] [CrossRef] [PubMed]
- Evanko, D. Hybridization chain reaction. Nat. Methods 2004, 1, 186–187. [Google Scholar] [CrossRef]
- Liu, S.F.; Wang, Y.; Ming, J.J.; Lin, Y.; Cheng, C.B.; Li, F. Enzyme-free and ultrasensitive electrochemical detection of nucleic acids by target catalyzed hairpin assembly followed with hybridization chain reaction. Biosens. Bioelectron. 2013, 49, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhou, H.; Zhu, W.P.; Li, H.B.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. Ultrasensitive electrochemical DNA detection based on dual amplification of circular strand-displacement polymerase reaction and hybridization chain reaction. Biosens. Bioelectron. 2013, 47, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Mayboroda, O.; Katakis, I.; O’Sullivan, C.K. Multiplexed isothermal nucleic acid amplification. Anal. Biochem 2018, 545, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J. Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, M.C.; Spoto, G. Integration of isothermal amplification methods in microfluidic devices: Recent advances. Biosens. Bioelectron. 2017, 90, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.U.; Nahar, S.; Safavieh, M.; Zourob, M. Real-time electrochemical detection of pathogen DNA using electrostatic interaction of a redox probe. Analyst 2013, 138, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.T.; Nadeau, J.G.; Spears, P.A.; Schram, J.L.; Nycz, C.M.; Shank, D.D. Multiplex strand displacement amplification (SDA) and detection of DNA sequences from Mycobacterium tuberculosis and other mycobacteria. Nucleic Acids Res. 1994, 22, 2670–2677. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Q.; Liu, Y.; Xin, C.; Zhao, J.K.; Liu, S.F. A cascade autocatalytic strand displacement amplification and hybridization chain reaction event for label-free and ultrasensitive electrochemical nucleic acid biosensing. Biosens. Bioelectron. 2018, 113, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rubab, M.; Shahbaz, H.M.; Olaimat, A.N.; Oh, D.H. Biosensors for rapid and sensitive detection of Staphylococcus aureus in food. Biosens. Bioelectron. 2018, 105, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Kim, K.H.; Deep, A. MOF-Bacteriophage Biosensor for Highly Sensitive and Specific Detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2017, 9, 33589–33598. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.Q.; Qiu, G.H.; Liang, Z.; Li, M.M.; Sun, B.; Qin, L.; Yang, S.P.; Chen, W.H.; Chen, J.X. A zinc(II)-based two-dimensional MOF for sensitive and selective sensing of HIV-1 ds-DNA sequences. Anal. Chim. Acta 2016, 922, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Han, J.; An, S.J.; Xie, G.; Chen, S.P. Ce(III, IV)-MOF electrocatalyst as signal-amplifying tag for sensitive electrochemical aptasensing. Biosens. Bioelectron. 2018, 109, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, B.; Ghani, M.; Shoushtari, A.M.; Rabiee, M. Electrochemical biosensors based on nanofibres for cardiac biomarker detection: A comprehensive review. Biosens. Bioelectron. 2016, 78, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Zeinhom, M.M.A.; Wang, Y.J.; Song, Y.; Zhu, M.J.; Lin, Y.H.; Du, D. A portable smart-phone device for rapid and sensitive detection of E. coli O157:H7 in Yoghurt and Egg. Biosens. Bioelectron. 2018, 99, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.I.; Shrivastava, S.; Duy, L.T.; Kim, B.Y.; Son, Y.M.; Lee, N.E. A smartphone imaging-based label-free and dual-wavelength fluorescent biosensor with high sensitivity and accuracy. Biosens. Bioelectron. 2017, 94, 643–650. [Google Scholar] [CrossRef] [PubMed]









| Pathogens | Gram | Virulence Factors | Epidemics | Food Source | Refs. |
|---|---|---|---|---|---|
| Salmonella | - | Enterotoxin | Typhoid fever, paratyphoid fever, gastroenteritis, and septicemia | Egg, raw milk and their products, commercial cold food dishes, raw poultry and meat | [3,4,5,6,7] |
| E. coli O157:H7 | - | Endotoxin, exotoxin, capsule, and adhesin. | Acute gastroenteritis and acute dysentery | Meat, fruits, vegetables, commercial cold food dishes, ready-to-eat food, drinking water | [7,8,9,10,11,12,13,14,15] |
| Listeria monocytogenes | + | Endogenous hormone, phagosome, and surface protein | Listeriosis | Frozen food, cheese, milk, meat products, ice, vegetable salad, ready-to-eat food, commercial cold food dishes | [7,16,17,18,19,20,21,22,23] |
| Staphylococcus aureus | + | Hemolytic toxin, leukocidin, enterotoxin, plasma coagulase, and deoxyribonuclease | Suppurative infection, pneumonia, pseudomembranous colitis, pericarditis, sepsis, septicemia | Milk, meat, eggs, fish and their products, commercial cold food dishes | [7,24,25,26] |
| Shigella | - | Endotoxin and exotoxin | Bacterial dysentery | Cooked food and raw material | [27,28,29] |
| Cronobacter | - | Enterotoxin, and adhesion factor | Necrotizing colitis, neonatal meningitis, and bacteremia | Powdered infant formula and milk powder | [30,31,32,33] |
| Vibrio parahemolyticus | - | Hemolysin and urease | Food poisoning, and acute diarrhea | Seafood such as fish, shrimp, crab, shellfish, and seaweed | [34,35,36] |
| Proteus | - | Endotoxin, and heat-resistant enterotoxin | Food poisoning, and acute diarrhea | Food of animal origin, bean products | [37,38] |
| Clostridium botulinum | + | Botulinum toxoid | Muscle relaxation paralysis, and respiratory paralysis | Canned products, cured meat | [39,40,41] |
| Bacillus cereus | + | Enterotoxin | Food poisoning | Leftovers of different meals, commercial cold food dishes | [7,42,43,44] |
| Campylobacter | - | Endotoxin, exotoxin, invasive protein, adhesion, and flagellum | Bacterial gastroenteritis | Raw chicken and by-products | [45,46] |
| Method | Derivative | Analysis Time | Advantages | Disadvantages | Refs. |
|---|---|---|---|---|---|
| Traditional microbiological culture | Chromogenic medium method | 5–7 days | High accuracy | Time-consuming, laborious, poor sensitivity and specificity | [56,57] |
| Immunological method | ELISA, immunomagnetic separation (IMS), immune colloidal gold technique (GICT) | 4 h | Rapid, relatively high sensitivity and specificity | High false positive rate and poor stability | [58,59,60,61,62,63] |
| PCR | Real time-PCR, multiple PCR | ≤2 h | Relatively sensitive and rapid, multiple detection | The need of expensive thermal cycle instruments and trained users | [64,65,66,67,68,69] |
| Nucleic acid-based isothermal amplification assays | LAMP, rolling circle amplification (RCA), saltatory rolling circle amplification (SRCA) | ≤2 h | No need for thermal cycle instruments, high sensitivity and selectivity | Not suitable for on-site detection | [70,71,72,73] |
| Biosensors | Based on signal amplification techniques such as nanotechnology | ≤2 h | Rapid, cost-effective, high sensitivity and selectivity | Most cannot achieve multiple detection | [74,75,76,77,78] |
| Methods | Principle | Evaluation |
|---|---|---|
| Adsorption | The skeleton of ssDNA is negatively charged, by modifying the surface of electrodes with positively charged substances or applying a positive potential, DNA can be absorbed on the electrodes. | Simple, with no need of any chemical reagents and DNA probes modification [101]. Low DNA hybridization efficiency. |
| Covalent binding | DNA is immobilized on the surface of electrodes through the formation of covalent bonds such as amide bonds, ester bonds, ether bonds, Au-S, and Ag-S et al. | Flexible structure, high efficiency of DNA immobilization and hybridization, but with the need of chemical reagents, and with the possibility of non-specific adsorption. |
| Affinity binding | Avidin is first adsorbed on the surface of the electrode by covalent binding or electrostatic adsorption, and then the biotin-modified DNA is immobilized on the electrode by affinity interaction between biotin and avidin. | The method is simple, stable and resistant to the extreme of temperature, pH, denatured detergents, and organic solvents [101]. |
| Electrodes | Targets | Detection Techniques | Linear Range | LOD | Ref. |
|---|---|---|---|---|---|
| Glassy carbon electrode (GCE) | Salmonella DNA | CV, EIS, DPV | 10–400 and 1–400 pM | 2.1 and 0.15 pM | [103] |
| Gold disk electrode | Salmonella typhimurium | CV, DPV | 102–108 CFU mL−1 | 3 CFU mL−1 | [104] |
| GCE | Staphylococcus aureus | CV, EIS | 10–106 CFU mL−1 | 10 CFU mL−1 | [107] |
| Gold electrode (GE) | Staphylococcus aureus | EIS | - | 10 CFU mL−1 | [108] |
| GE | Escherichia coli, K. pneumoniae | EIS | 102–106 CFU mL−1 | 100 CFU mL−1 | [109] |
| GE | E. faecalis, B. subtilis | EIS | 103–106 CFU mL−1 | 1000 CFU mL−1 | [109] |
| GE | S. aureus, E. faecalis, P. aeruginosa, E. coli and Salmonella typhimurium | CV, EIS | 101–104 CFU mL−1 | 10 CFU mL−1 | [110] |
| Indium tin oxide (ITO) | Salmonella typhimurium DNA | CV, DPV | 10 fM–50 nM | 10 fM | [111] |
| ITO | Escherichia coli O157:H7 DNA | CV, EIS | 1 uM–10 fM | 10 fM | [112] |
| GE | Bacillus cereus spore simulant | EIS | 104–5 × 106 CFU mL−1 | 3000 CFU mL−1 | [113] |
| Carbon paste electrode (CPE) | Aeromonas hydrophila DNA | SWV | - | 160 fM | [114] |
| Carbon ionic liquid electrode (CILE) | Listeria monocytogenes DNA | CV, EIS, DPV | 1 uM–1 pM | 290 fM | [85] |
| Pt/Ir electrodes | Listeria monocytogenes | CV, DPV | - | 100 CFU mL−1 | [115] |
| ITO | Salmonella typhimurium DNA | DPV, EIS | 4 aM–24 fM | 4 aM | [116] |
| GE | Enterobacteriaceae bacteria DNA | SWV, DPV, EIS | 0.01 pM–1 nM | 8.7 fM | [117] |
| GE | Salmonella | SWV, DPV, EIS | 2 × 102–2 × 106 CFU mL−1 | 200 CFU mL−1 | [118] |
| GE | Bacillus subtilis DNA | DPV | 0.1 fM-20 fM | 0.08 fM | [119] |
| GCE | Salmonella | CV, EIS | 75-7.5 × 105 CFU mL−1 | 25 CFU mL−1 | [120] |
| ITO | Salmonella typhimurium | CV, EIS | - | 10 CFU mL−1 | [121] |
| Pencil graphite electrode (PGE) | Bacillus cereus | DPV, EIS | 100–107 CFU mL−1 | 9.4 pM | [76] |
| CILE | Yersinia enterocolitica DNA | DPV | 1 uM–10 PM | 1.76 pM | [122] |
| GCE | E. coli O157:H7 DNA | CV, EIS, DPV | - | 19.7 fM | [79] |
| GE | Salmonella typhimurium | DPV | 72–7.2 × 106 CFU mL−1 | 28 CFU mL−1 | [123] |
| GCE | DNA | ASV, EIS | - | 100 aM | [124] |
| Redox Active Molecule | Classification | Target | Principle | Refs. |
|---|---|---|---|---|
| Methylene Blue (MB) | Organic dye | Bacillus cereus; Listeria monocytogenes | MB covalently interacts with G bases of DNA | [76,85] |
| Toluidine Blue (TB) | Organic dye | Enterococcus faecalis | TB binds to a negatively charged phosphate group | [80] |
| Oracet Blue (OB) | Organic dye | Helicobacter pylori | The hydrophobic rigid plane of OB inserts into the dsDNA base pair | [128,129,131] |
| Hoechst 33258 | Organic dye | Aeromona hydrophila | Hoechst 33258 can bind to dsDNA by minor and major groove interaction | [114] |
| [Ru(phen)3]2+ | Metal complex | Aeromona hydrophila | Ru(phen)32+ can intercalate into the groove of dsDNA | [97,129] |
| Daunomycin | Drug molecular | Aeromona hydrophila | The molecular carbocyclic moiety can be inserted into the base pair of the DNA helix, and the amino sugar moiety generate electrostatic interaction with the phosphate backbone of the DNA | [114] |
| Nanocomposites/Electrode | Features | Immobilizing Methods of DNA | Targets | LOD (mol/L) | Ref. |
|---|---|---|---|---|---|
| AgNCs/AuNPs/GCE | AgNCs are used as direct signal indicator and AuNPs as carrier for signal amplification | By the Au-S bonds between AuNPs and SH-DNA | Salmonella | 1.62 × 10−16 | [142] |
| CTS/V2O5/MWCN/CILE | Great biocompatibility of V2O5 nanobelt and excellent electron transfer ability of MWCNTs | CTS can be used for DNA immobilization by electrostatic attraction | Yersinia enterocolitica | 1.76 × 10−12 | [122] |
| NiO/GR/CILE | Graphene and nickel oxide composite possess high surface area and strong affinity with phosphate groups of ssDNA | By the strong affinity between NiO and phosphate groups of ssDNA | Salmonella enteritidis | 3.12 × 10−14 | [143] |
| DpAu/GOx/GCE | GOx has fast electron transfer kinetics and large specific surface area. Thi has good electrochemical redox active properties. Au@SiO2 can provide a microenvironment to retain the DNA tag conformation and make them free in orientation | By the Au-S bonds between Au@SiO2 and SH-DNA | E.coli O157:H7 | 1.0 × 10−11 | [144] |
| Au/GR/CILE | Graphene (GR) possesses high thermal conductivity, good mechanical strength, high mobility of charge carriers, big specific surface area and upstanding electrical properties. The dendritic nanogold provides more sites for the self-assembly of MAA on the electrode surface | By the covalent bonds between the amine groups of ssDNA and the carboxyl group modified on the CILE surface | Listeria monocyto | 2.9 × 10−13 | [85] |
| CTS/Co3O4/GR/CILE | The nanocomposite film has a very large surface area, good conductivity and excellent porous structure, which lead to the measurable currents even for low concentrations of ssDNA sequence | ssDNA was immobilized on the CTS/Co3O4/GR/CILE surface by electrostatic attraction | Staphylococcus aureus | 4.3 × 10−13 | [145] |
| AuNPs/CS/MWCNT/AuE | CS–MWCNTs greatly increase effective surface area and electron conductivity. AuNPs provide a biocompatible interface for DNA | By the Au-S bonds between AuNPs and SH-DNA | Staphylococcus aureus | 3.3 × 10−16 | [141] |
| CeO2NPs/RGO/GCE | RGO has an extremely large surface area, excellent thermal and electrical conductivity; CeO2 possesses high catalytic activity and biocompatibility | By the Π-Π stacking between RGO and DNA bases and electrostatic attraction between CeO2NPs and DNA | Aeromonas hydrophila | 1.0 × 10−16 | [140] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Zhang, Y.; Yang, Q.; Yuan, N.; Zhang, W. Review of Electrochemical DNA Biosensors for Detecting Food Borne Pathogens. Sensors 2019, 19, 4916. https://doi.org/10.3390/s19224916
Wu Q, Zhang Y, Yang Q, Yuan N, Zhang W. Review of Electrochemical DNA Biosensors for Detecting Food Borne Pathogens. Sensors. 2019; 19(22):4916. https://doi.org/10.3390/s19224916
Chicago/Turabian StyleWu, Qiaoyun, Yunzhe Zhang, Qian Yang, Ning Yuan, and Wei Zhang. 2019. "Review of Electrochemical DNA Biosensors for Detecting Food Borne Pathogens" Sensors 19, no. 22: 4916. https://doi.org/10.3390/s19224916
APA StyleWu, Q., Zhang, Y., Yang, Q., Yuan, N., & Zhang, W. (2019). Review of Electrochemical DNA Biosensors for Detecting Food Borne Pathogens. Sensors, 19(22), 4916. https://doi.org/10.3390/s19224916




