Detection of the Plant Pathogen Pseudomonas Syringae pv. Lachrymans on Antibody-Modified Gold Electrodes by Electrochemical Impedance Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Bacterial Samples Preparation
2.3. Bacterial Samples Verification and Quantification
2.4. Apparatus
2.5. Electrochemical Measurements
2.6. Preparation of Electrodes
2.7. Fabrication of Electrochemical Immunosensor
3. Results
3.1. The Sensing Principle of Biosensor
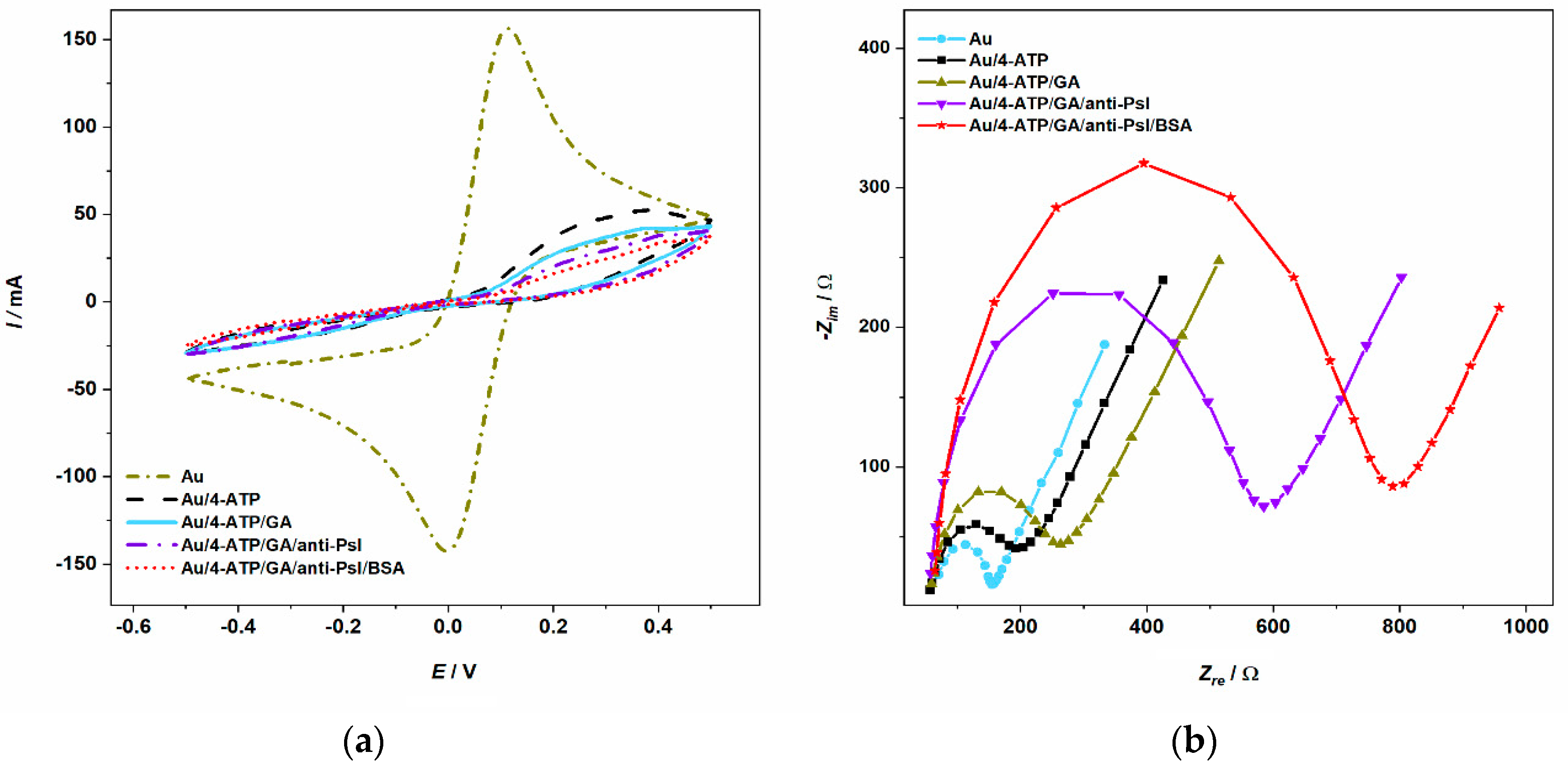
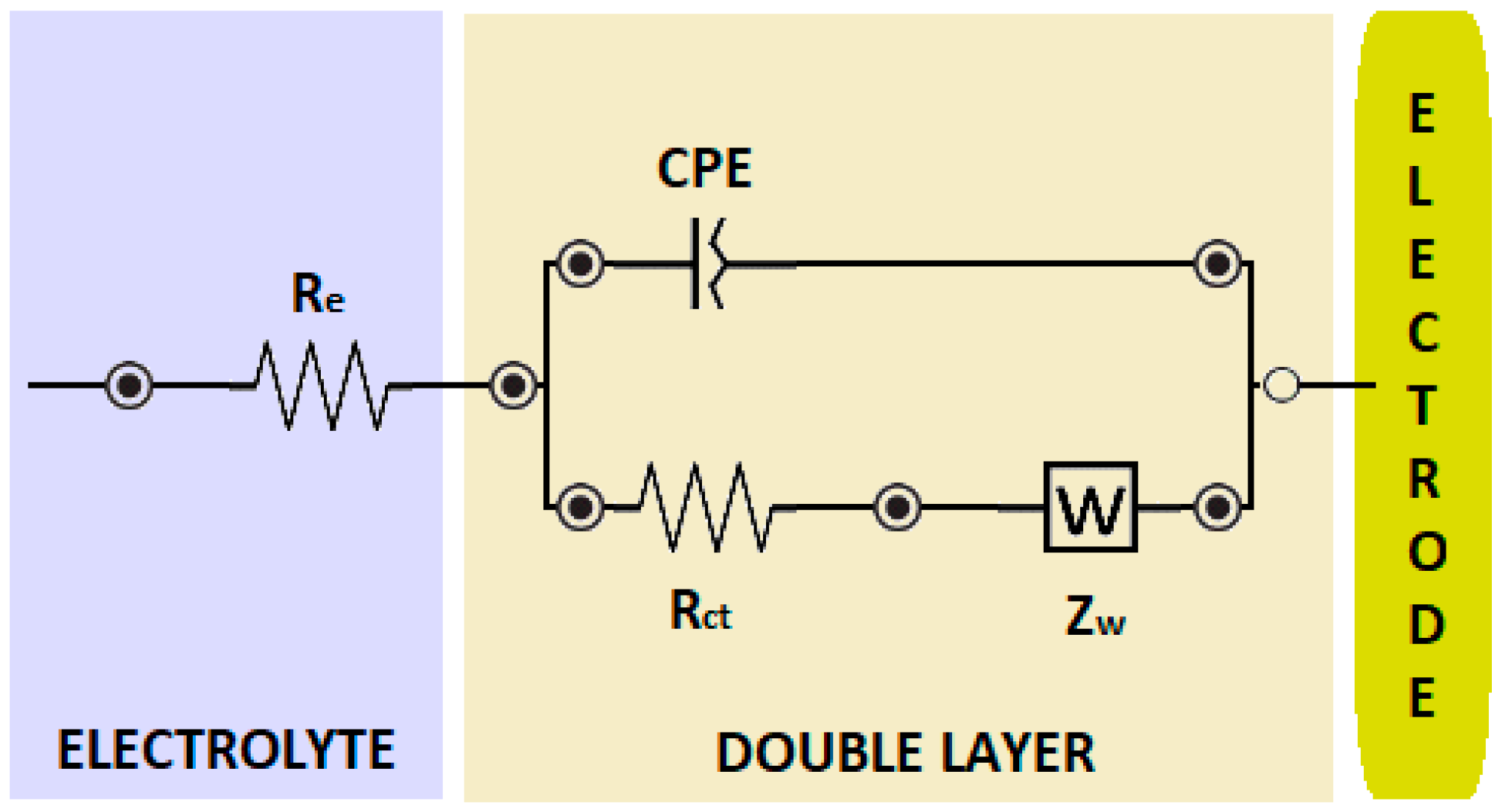
3.2. Electrochemical Characterization of Biosensor
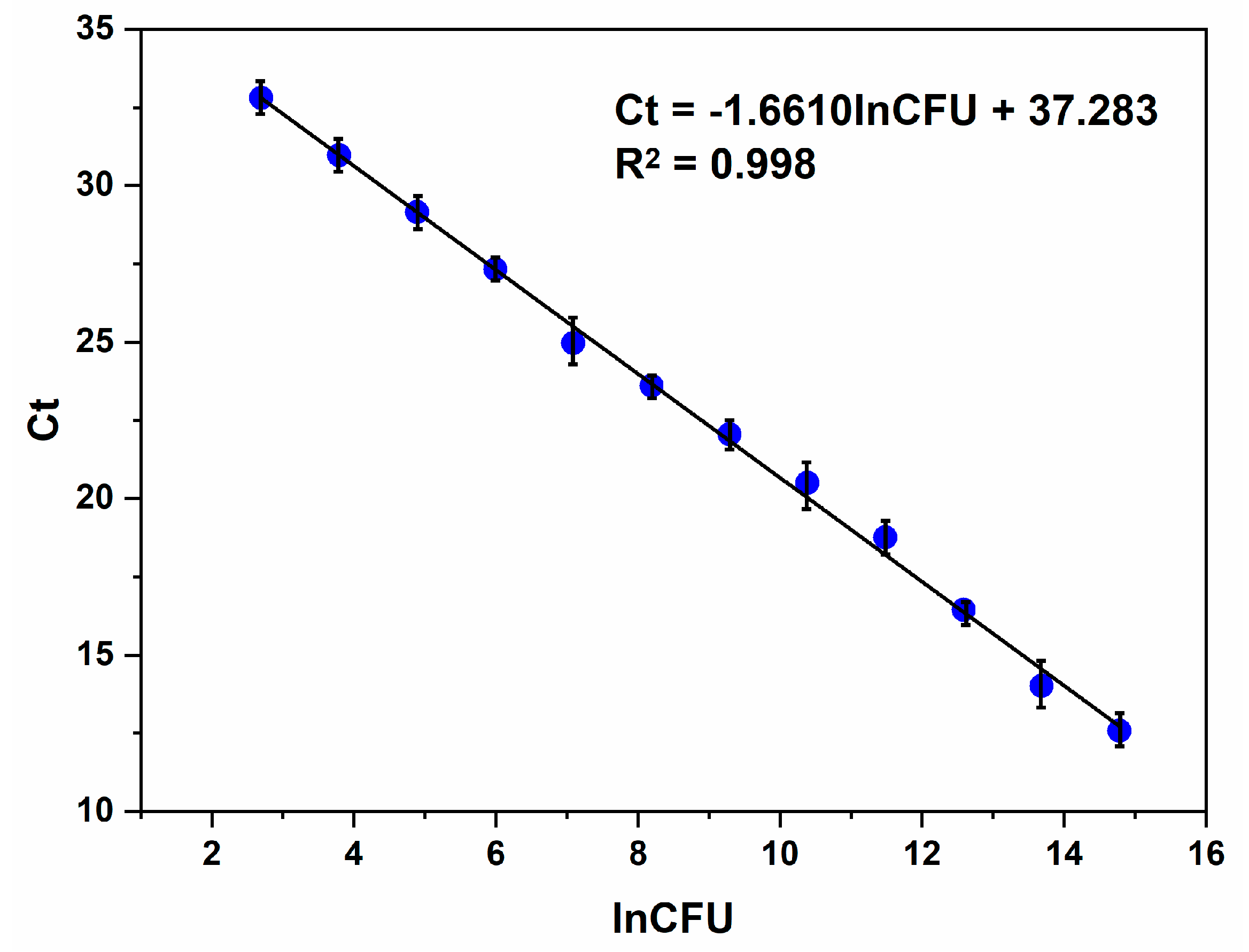
3.3. Psl Bacteria Identification by qPCR
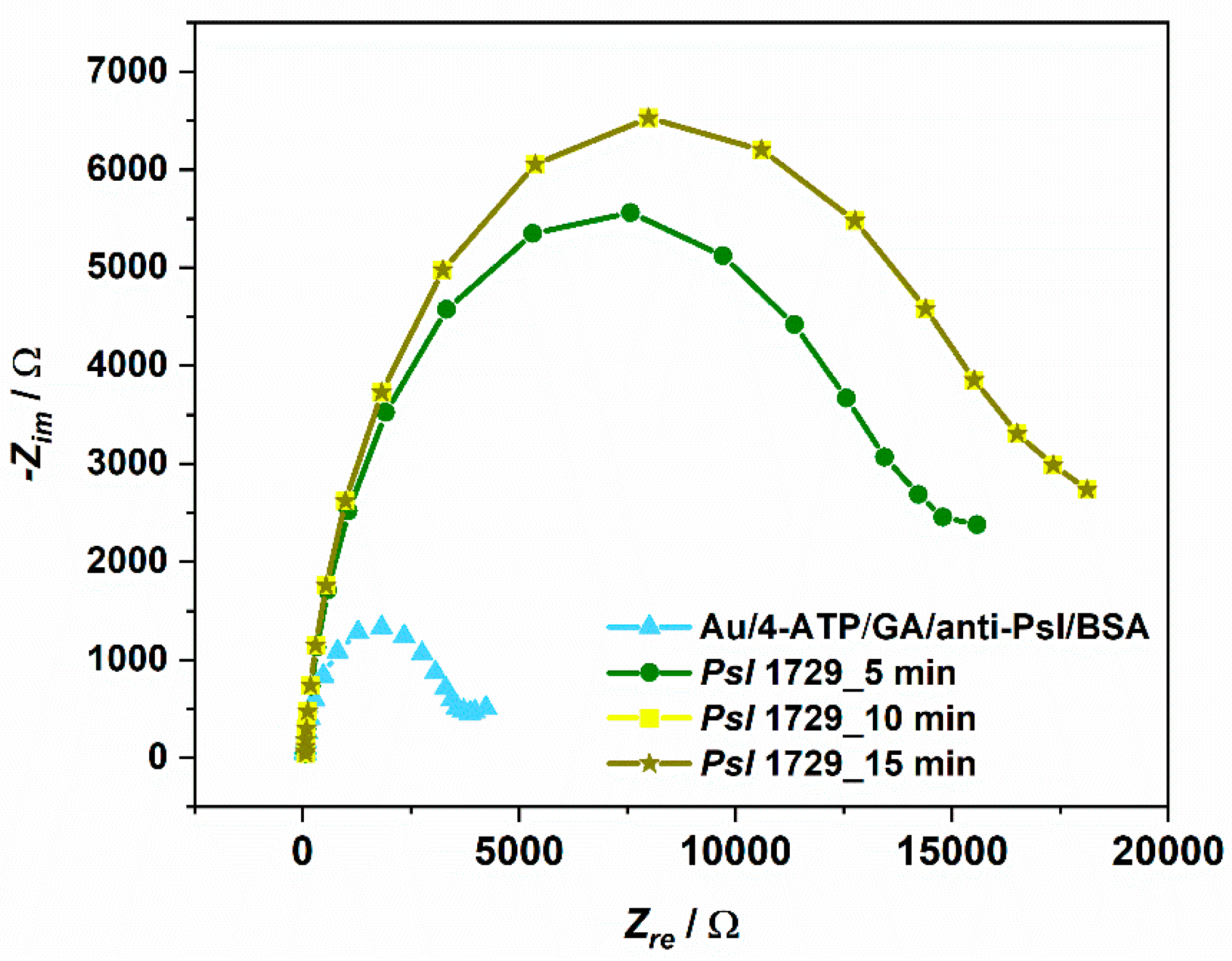
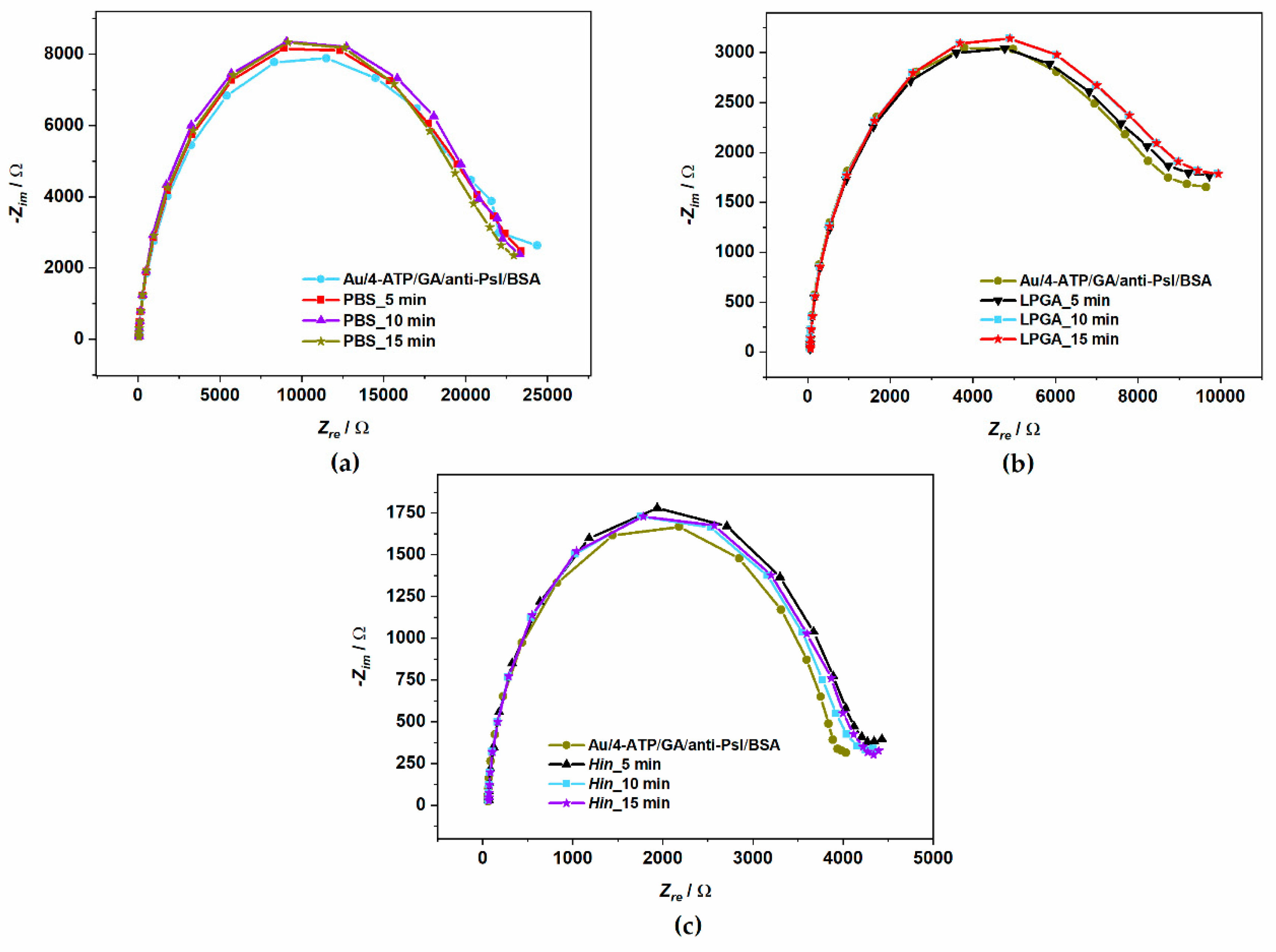
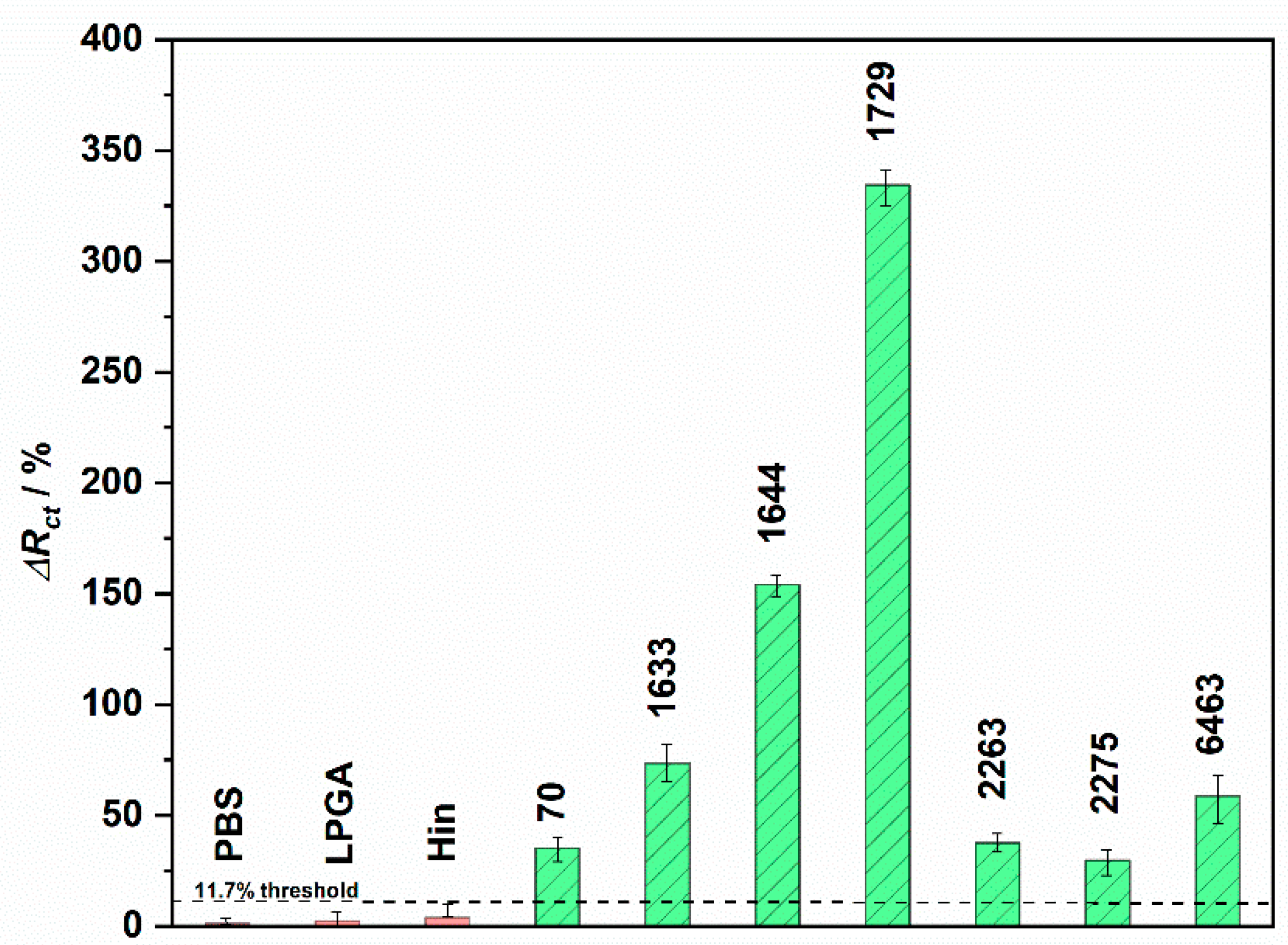
3.4. Psl Bacteria Detection on Developed Electrochemical Immunosensor
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Young, J.M. Taxonomy of pseudomonas syringae. J. Plant Pathol. 2010, 92, S5–S14. [Google Scholar]
- Baltrus, D.A.; McCann, H.C.; Guttman, D.S. Evolution, genomics and epidemiology of Pseudomonas syringae: Challenges in Bacterial Molecular Plant Pathology. Mol. Plant Pathol. 2017, 18, 152–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, M.S.H.; Morgan, R.L.; Sarkar, S.F.; Wang, P.W.; Guttman, D.S. Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 2005, 71, 5182–5191. [Google Scholar] [CrossRef] [Green Version]
- Horst, R.K. Westcott’s Plant Disease Handbook, 7th ed.; Springer Netherlands: Dordrecht, The Netherlands, 2008; ISBN 978-1-4020-4585-1. [Google Scholar]
- Słomnicka, R.; Olczak-Woltman, H.; Korzeniewska, A.; Gozdowski, D.; Niemirowicz-Szczytt, K.; Bartoszewski, G. Genetic mapping of psl locus and quantitative trait loci for angular leaf spot resistance in cucumber (Cucumis sativus L.). Mol. Breed. 2018, 38, 111. [Google Scholar] [CrossRef] [Green Version]
- Skłodowska, M.; Gajewska, E.; Naliwajski, M.; Wielanek, M.; Kuźniak, E.; Chojak-Koźniewska, J. Benzothiadiazole and nitrogen source modify the nitrogen metabolism in cucumber infected with Pseudomonas syringae pv. lachrymans. Sci. Hortic. 2019, 246, 289–297. [Google Scholar] [CrossRef]
- Kuźniak, E.; Wielanek, M.; Chwatko, G.; Głowacki, R.; Libik-Konieczny, M.; Piątek, M.; Gajewska, E.; Skłodowska, M. Salicylic acid and cysteine contribute to arbutin-induced alleviation of angular leaf spot disease development in cucumber. J. Plant Physiol. 2015, 181, 9–13. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Messéan, A.; Morris, C.E. Insights into epidemiology and control of diseases of annual plants caused by the Pseudomonas syringae species complex. J. Gen. Plant Pathol. 2015, 81, 331–350. [Google Scholar] [CrossRef]
- Harighi, B. Angular Leaf Spot of Cucumber Caused by Pseudomonas syringae pv. lachrymans in Kurdistan. Plant Dis. 2007, 91, 769. [Google Scholar] [CrossRef]
- Słomnicka, R.; Olczak-Woltman, H.; Oskiera, M.; Schollenberger, M.; Niemirowicz-Szczytt, K.; Bartoszewski, G. Genome analysis of Pseudomonas syringae pv. lachrymans strain 814/98 indicates diversity within the pathovar. Eur. J. Plant Pathol. 2018, 151, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Olczak-Woltman, H.; Schollenberger, M.; Mądry, W.; Niemirowicz-Szczytt, K. Evaluation of cucumber (Cucumis sativus) cultivars grown in Eastern Europe and progress in breeding for resistance to angular leaf spot (Pseudomonas syringae pv. lachrymans). Eur. J. Plant Pathol. 2008, 122, 385–393. [Google Scholar] [CrossRef]
- Kůdela, V.; Lebeda, A. Response of wild Cucumis species to inoculation with Pseudomonas syringae pv. lachrymans. Genet. Resour. Crop. Evol. 1997, 44, 271–275. [Google Scholar] [CrossRef]
- Plantwise Knowledge Bank. Available online: https://www.plantwise.org/KnowledgeBank/pmdg/20197800204 (accessed on 13 September 2019).
- Słomnicka, R.; Olczak-Woltman, H.; Bartoszewski, G.; Niemirowicz-Szczytt, K. Genetic and pathogenic diversity of Pseudomonas syringae strains isolated from cucurbits. Eur. J. Plant Pathol. 2015, 141, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Alleyne, A. Identification of Pseudomonas syringae pv. lachrymans in Barbados by rep-PCR. J. Agric. Sci. Techol. B 2011, 8, 593–597. [Google Scholar]
- Ruinelli, M.; Blom, J.; Smits, T.H.M.; Pothier, J.F. Comparative genomics and pathogenicity potential of members of the Pseudomonas syringae species complex on Prunus spp. BMC Genom. 2019, 20, 172. [Google Scholar] [CrossRef]
- Vasebi, Y.; Khakvar, R.; Faghihi, M.M.; Vinatzer, B.A. Genomic and pathogenic properties of Pseudomonas syringae pv. syringae strains isolated from apricot in East Azerbaijan province, Iran. Biocatal. Agric. Biotechnol. 2019, 19, 101167. [Google Scholar] [CrossRef]
- González, A.J.; Rodicio, M.R.; Mendoza, M.C. Identification of an Emergent and Atypical Pseudomonas viridiflava Lineage Causing Bacteriosis in Plants of Agronomic Importance in a Spanish Region. Appl. Environ. Microbiol. 2003, 69, 2936–2941. [Google Scholar] [CrossRef] [Green Version]
- Cimmino, A.; Iannaccone, M.; Petriccione, M.; Masi, M.; Evidente, M.; Capparelli, R.; Scortichini, M.; Evidente, A. An ELISA method to identify the phytotoxic Pseudomonas syringae pv. actinidiae exopolysaccharides: A tool for rapid immunochemical detection of kiwifruit bacterial canker. Phytochem. Lett. 2017, 19, 136–140. [Google Scholar] [CrossRef]
- Wang, W.; Feng, M.; Kong, D.; Liu, L.; Song, S.; Xu, C. Development of an immunochromatographic strip for the rapid detection of Pseudomonas syringae pv. maculicola in broccoli and radish seeds. Food Agric. Immunol. 2015, 26, 738–745. [Google Scholar] [CrossRef]
- Casano, F.J.; Hung, S.Y.; Wells, J.M. Differentiation of some pathovars of Pseudomonas syringae with monoclonal antibodies1. EPPO Bull. 1987, 17, 173–176. [Google Scholar] [CrossRef]
- Cho, M.S.; Jeon, Y.H.; Kang, M.J.; Ahn, H.I.; Baek, H.J.; Na, Y.W.; Choi, Y.M.; San Kim, T.; Park, D.S. Sensitive and specific detection of phaseolotoxigenic and nontoxigenic strains of Pseudomonas syringae pv. phaseolicola by TaqMan real-time PCR using site-specific recombinase gene sequences. Microbiol. Res. 2010, 165, 565–572. [Google Scholar]
- Manceau, C.; Horvais, A. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl. Environ. Microbiol. 1997, 63, 498–505. [Google Scholar]
- Bereswill, S.; Bugert, P.; Völksch, B.; Ullrich, M.; Bender, C.L.; Geider, K. Identification and relatedness of coronatine-producing Pseudomonas syringae pathovars by PCR analysis and sequence determination of the amplification products. Appl. Environ. Microbiol. 1994, 60, 2924–2930. [Google Scholar]
- Rees-George, J.; Vanneste, J.L.; Cornish, D.A.; Pushparajah, I.P.S.; Yu, J.; Templeton, M.D.; Everett, K.R. Detection of Pseudomonas syringae pv. actinidiae using polymerase chain reaction (PCR) primers based on the 16S–23S rDNA intertranscribed spacer region and comparison with PCR primers based on other gene regions. Plant Pathol. 2010, 59, 453–464. [Google Scholar] [CrossRef]
- Sarkar, S.F.; Guttman, D.S. Evolution of the Core Genome of Pseudomonas syringae, a Highly Clonal, Endemic Plant Pathogen. Appl. Environ. Microbiol. 2004, 70, 1999–2012. [Google Scholar] [CrossRef] [Green Version]
- Gazdik, F.; Penazova, E.; Cechova, J.; Baranek, M.; Eichmeier, A. Quantitative real-time PCR assay for rapid detection of Pseudomonas amygdali pv. lachrymans in cucumber leaf rinse. J. Plant Dis. Prot. 2019, 126, 517–528. [Google Scholar] [CrossRef]
- Meng, X.-L.; Xie, X.-W.; Shi, Y.-X.; Chai, A.-L.; Ma, Z.-H.; Li, B.-J. Evaluation of a loop-mediated isothermal amplification assay based on hrpZ gene for rapid detection and identification of Pseudomonas syringae pv. lachrymans in cucumber leaves. J. Appl. Microbiol. 2017, 122, 441–449. [Google Scholar] [CrossRef] [Green Version]
- Fogliano, V.; Gallo, M.; Vinale, F.; Ritieni, A.; Randazzo, G.; Greco, M.; Lops, R.; Graniti, A. Immunological detection of syringopeptins produced by Pseudomonas syringae pv. lachrymans. Physiol. Mol. Plant. Pathol. 1999, 55, 255–261. [Google Scholar] [CrossRef]
- Vizzini, P.; Braidot, M.; Vidic, J.; Manzano, M. Electrochemical and Optical Biosensors for the Detection of Campylobacter and Listeria: An Update Look. Micromachines 2019, 10, 500. [Google Scholar] [CrossRef] [Green Version]
- Khater, M.; de la Escosura-Muñiz, A.; Merkoçi, A. Biosensors for plant pathogen detection. Biosens. Bioelectron. 2017, 93, 72–86. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, B.; Kalyani, N.; Das, S.; Anand, A.; Sharma, T.K. Chapter 2—Nano-realm for point-of-care (POC) bacterial diagnostics. In Methods in Microbiology; Gurtler, V., Ball, A.S., Soni, S., Eds.; Nanotechnology; Academic Press: Cambridge, MA, USA, 2019; Volume 46, pp. 19–42. [Google Scholar]
- Jarocka, U.; Radecka, H.; Malinowski, T.; Michalczuk, L.; Radecki, J. Detection of Prunus Necrotic Ringspot Virus in Plant Extracts with Impedimetric Immunosensor based on Glassy Carbon Electrode. Electroanalysis 2013, 25, 433–438. [Google Scholar] [CrossRef]
- Malecka, K.; Michalczuk, L.; Radecka, H.; Radecki, J. Ion-Channel Genosensor for the Detection of Specific DNA Sequences Derived from Plum Pox Virus in Plant Extracts. Sensors 2014, 14, 18611–18624. [Google Scholar] [CrossRef] [Green Version]
- Drygin, Y.F.; Blintsov, A.N.; Grigorenko, V.G.; Andreeva, I.P.; Osipov, A.P.; Varitzev, Y.A.; Uskov, A.I.; Kravchenko, D.V.; Atabekov, J.G. Highly sensitive field test lateral flow immunodiagnostics of PVX infection. Appl. Microbiol. Biotechnol. 2012, 93, 179–189. [Google Scholar] [CrossRef]
- Zhao, W.; Lu, J.; Ma, W.; Xu, C.; Kuang, H.; Zhu, S. Rapid on-site detection of Acidovorax avenae subsp. citrulli by gold-labeled DNA strip sensor. Biosens. Bioelectron. 2011, 26, 4241–4244. [Google Scholar] [CrossRef]
- Vaseghi, A.; Safaie, N.; Bakhshinejad, B.; Mohsenifar, A.; Sadeghizadeh, M. Detection of Pseudomonas syringae pathovars by thiol-linked DNA–Gold nanoparticle probes. Sens. Actuators B-Chem. 2013, 181, 644–651. [Google Scholar] [CrossRef]
- Lau, H.Y.; Wu, H.; Wee, E.J.H.; Trau, M.; Wang, Y.; Botella, J.R. Specific and Sensitive Isothermal Electrochemical Biosensor for Plant Pathogen DNA Detection with Colloidal Gold Nanoparticles as Probes. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Cinti, S.; Volpe, G.; Piermarini, S.; Delibato, E.; Palleschi, G. Electrochemical Biosensors for Rapid Detection of Foodborne Salmonella: A Critical Overview. Sensors 2017, 17, 1910. [Google Scholar] [CrossRef]
- Amouzadeh Tabrizi, M.; Shamsipur, M. A label-free electrochemical DNA biosensor based on covalent immobilization of salmonella DNA sequences on the nanoporous glassy carbon electrode. Biosens. Bioelectron. 2015, 69, 100–105. [Google Scholar] [CrossRef]
- Felix, F.S.; Angnes, L. Electrochemical immunosensors—A powerful tool for analytical applications. Biosens. Bioelectron. 2018, 102, 470–478. [Google Scholar] [CrossRef]
- Catanante, G.; Rhouati, A.; Hayat, A.; Marty, J.L. An Overview of Recent Electrochemical Immunosensing Strategies for Mycotoxins Detection. Electroanalysis 2016, 28, 1750–1763. [Google Scholar] [CrossRef]
- Grabowska, I.; Malecka, K.; Jarocka, U.; Radecki, J.; Radecka, H. Electrochemical biosensors for detection of avian influenza virus--current status and future trends. Acta Biochim. Pol. 2014, 61, 471–478. [Google Scholar] [CrossRef] [Green Version]
- Bondarenko, A.S.; Ragoisha, G.A. Progress in Chemometrics Research; Pomerantsev, A.L., Ed.; Nova Science Publishers: New York, NY, USA, 2005; pp. 89–102. Available online: http://www.abc.chemistry.bsu.by/vi/analyser/ (accessed on 13 September 2019).
- Fowler, J.M.; Wong, D.K.Y.; Halsall, H.B.; Heineman, W.R. Chapter 5—Recent developments in electrochemical immunoassays and immunosensors. In Electrochemical Sensors, Biosensors and Their Biomedical Applications; Zhang, X., Ju, H., Wang, J., Eds.; Academic Press: San Diego, CA, USA, 2008; pp. 115–143. ISBN 978-0-12-373738-0. [Google Scholar]
- Tsugimura, K.; Ohnuki, H.; Wu, H.; Endo, H.; Tsuya, D.; Izumi, M. Oriented antibody immobilization on self-assembled monolayers applied as impedance biosensors. J. Phys. Conf. Ser. 2017, 924, 012015. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Fu, Y.; Fang, W.; Li, Y. Electrochemical Impedance Immunosensor Based on Self-Assembled Monolayers for Rapid Detection of Escherichia coli O157:H7 with Signal Amplification Using Lectin. Sensors 2015, 15, 19212–19224. [Google Scholar] [CrossRef]


| Scientific Name. | Collection No. | Geographic Origin |
|---|---|---|
| Pseudomonas syringae pv. lachrymans | 70 | France |
| Pseudomonas syringae pv. lachrymans | 1633 | Canada |
| Pseudomonas syringae pv. lachrymans | 1644 | Hungary |
| Pseudomonas syringae pv. lachrymans | 1729 | Denmark |
| Pseudomonas syringae pv. lachrymans | 2263 | Romania |
| Pseudomonas syringae pv. lachrymans | 2275 | Romania |
| Pseudomonas syringae pv. lachrymans | 6463 | Hungary |
| Modification Step | Re [Ω] | Rct [Ω] | CPE [µFΩ−1sn] | n * |
|---|---|---|---|---|
| Au | 64.5 | 83.6 | 6.25 | 1.007 |
| Au/4-ATP | 54.3 | 138.4 | 6.61 | 0.850 |
| Au/4-ATP/GA | 55.0 | 194.0 | 3.14 | 0.889 |
| Au/4-ATP/GA/anti-Psl | 55.0 | 494.8 | 1.19 | 0.949 |
| Au/4-ATP/GA/anti-Psl/BSA | 62.1 | 679.4 | 1.08 | 0.954 |
| Sample | Ct | CFU/mL | Test Result |
|---|---|---|---|
| Psl 70 | 14.07 | 1172325569 | positive |
| Psl 1633 | 15.07 | 642100427.8 | positive |
| Psl 1644 | 14.87 | 724255855.1 | positive |
| Psl 1729 | 15.06 | 634377828.5 | positive |
| Psl 2263 | 15.59 | 469488930.4 | positive |
| Psl 2275 | 15.31 | 555708969.5 | positive |
| Psl 6463 | 15.09 | 645964611.3 | positive |
| 17 ng DNA (positive control). | 10.21 | 2629019000 | positive |
| PBS | - | - | negative |
| LPGA medium | - | - | negative |
| Haemophilus influenzae | - | - | negative |
| Method | Target Analyte | LOD | Range of Determination | Year | Ref |
|---|---|---|---|---|---|
| Real-time qPCR | Bacterial DNA | 260 CFU/mL | 271–6.94 × 107 CFU/mL | 2019 | [27] |
| LAMP | Bacterial cells | 104 CFU/mL | 104–108 CFU/mL | 2017 | [28] |
| ELISA | Syringopeptins | 0 ± 05 µg/well | 0 ± 1–10 µg | 1999 | [29] |
| Colorimetric | Bacterial DNA | 15 ng/μL | 90–200 ng/L | 2013 | [37] |
| RPA + DPV | Bacterial DNA | 21.4 pM | 21.4 pM–2.140 µM | 2017 | [38] |
| EIS. | Bacterial cells | 337 CFU/mL | 1 × 103–1.2 × 105 CFU/mL | 2019 | This work |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cebula, Z.; Żołędowska, S.; Dziąbowska, K.; Skwarecka, M.; Malinowska, N.; Białobrzeska, W.; Czaczyk, E.; Siuzdak, K.; Sawczak, M.; Bogdanowicz, R.; et al. Detection of the Plant Pathogen Pseudomonas Syringae pv. Lachrymans on Antibody-Modified Gold Electrodes by Electrochemical Impedance Spectroscopy. Sensors 2019, 19, 5411. https://doi.org/10.3390/s19245411
Cebula Z, Żołędowska S, Dziąbowska K, Skwarecka M, Malinowska N, Białobrzeska W, Czaczyk E, Siuzdak K, Sawczak M, Bogdanowicz R, et al. Detection of the Plant Pathogen Pseudomonas Syringae pv. Lachrymans on Antibody-Modified Gold Electrodes by Electrochemical Impedance Spectroscopy. Sensors. 2019; 19(24):5411. https://doi.org/10.3390/s19245411
Chicago/Turabian StyleCebula, Zofia, Sabina Żołędowska, Karolina Dziąbowska, Marta Skwarecka, Natalia Malinowska, Wioleta Białobrzeska, Elżbieta Czaczyk, Katarzyna Siuzdak, Mirosław Sawczak, Robert Bogdanowicz, and et al. 2019. "Detection of the Plant Pathogen Pseudomonas Syringae pv. Lachrymans on Antibody-Modified Gold Electrodes by Electrochemical Impedance Spectroscopy" Sensors 19, no. 24: 5411. https://doi.org/10.3390/s19245411
APA StyleCebula, Z., Żołędowska, S., Dziąbowska, K., Skwarecka, M., Malinowska, N., Białobrzeska, W., Czaczyk, E., Siuzdak, K., Sawczak, M., Bogdanowicz, R., & Nidzworski, D. (2019). Detection of the Plant Pathogen Pseudomonas Syringae pv. Lachrymans on Antibody-Modified Gold Electrodes by Electrochemical Impedance Spectroscopy. Sensors, 19(24), 5411. https://doi.org/10.3390/s19245411







