Fluorescent Method for the Detection of Biothiols Using an Ag+-Mediated Conformational Switch
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Apparatus
2.3. Optimization of the Experimental Conditions
2.4. Fluorescence Detection of Glutathione and Cysteine
2.5. Selectivity Assay
2.6. Sample Assay
3. Results and Discussion
3.1. Principle of the Detection of Biothiols
3.2. Method Feasibility
3.3. Optimization of Experimental Conditions
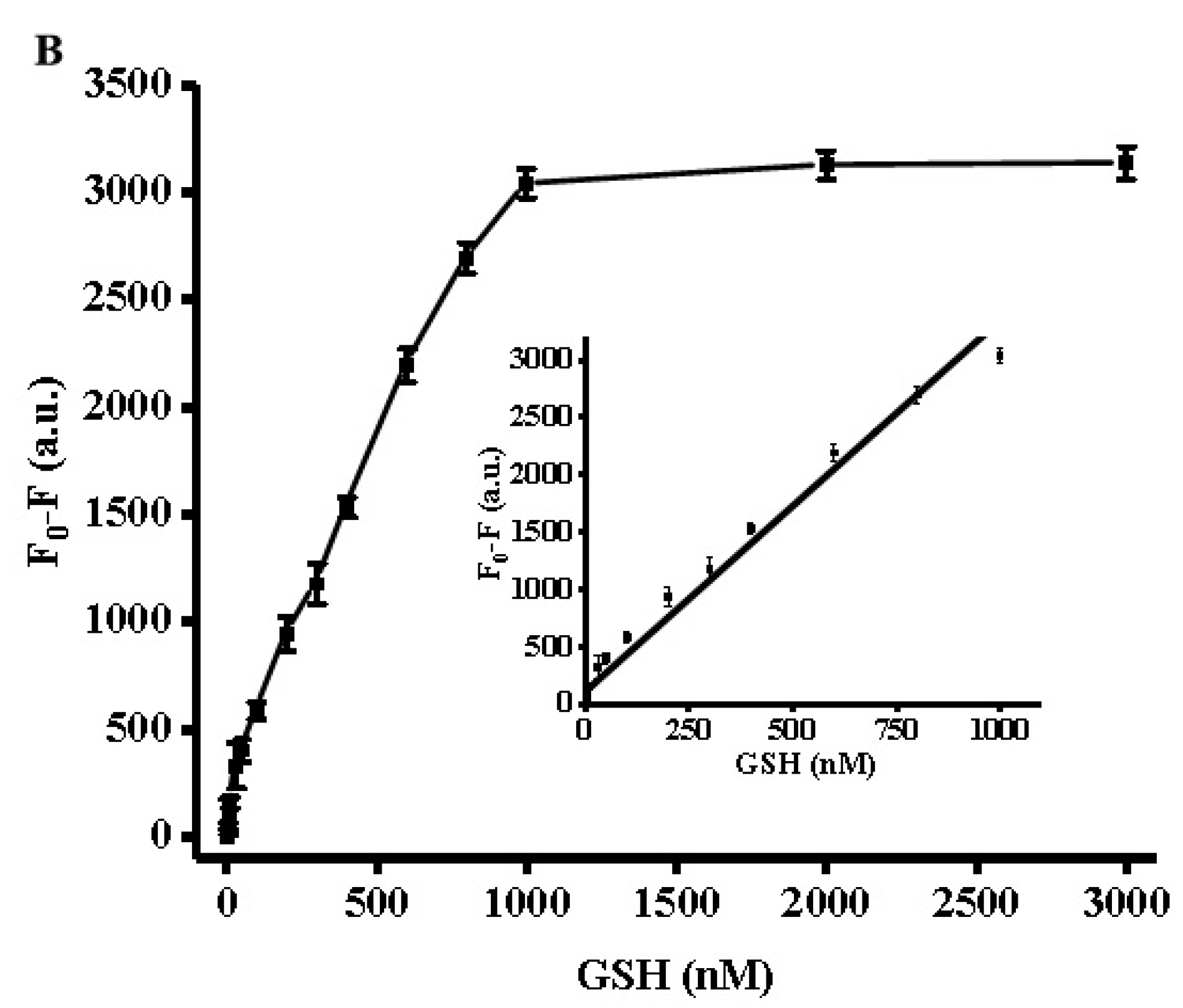
3.4. Quantification of Biothiols Activity
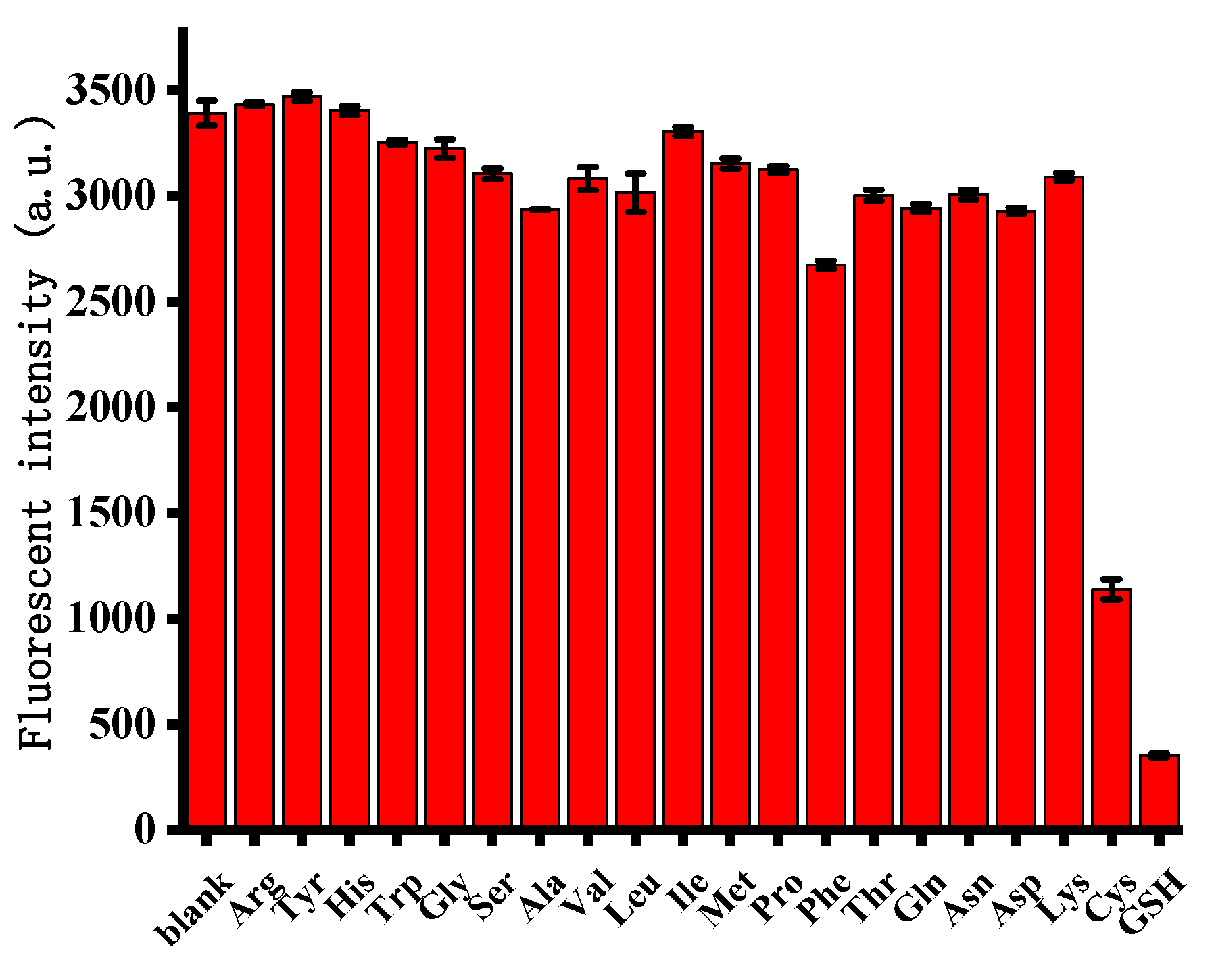
3.5. Selectivity for Cysteine and Glutathione
3.6. Detection of Biothiols in Serum Samples
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yang, X.; Guo, Y.; Strongin, R.M. Conjugate Addition/Cyclization Sequence Enables Selective and Simultaneous Fluorescence Detection of Cysteine and Homocysteine. Angew. Chem. Int. Ed. 2011, 50, 10690–10693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, B.; Wang, E. Oligonucleotide-stabilized fluorescent silver nanoclusters for sensitive detection of biothiols in biological fluids. Biosens. Bioelectron. 2011, 26, 2585–2589. [Google Scholar] [CrossRef] [PubMed]
- Scholz, R.W.; Graham, K.S.; Gumpricht, E.; Reddy, C.C. Mechanism of Interaction of Vitamin E and Glutathione in the Protection against Membrane Lipid Peroxidation. Ann. N. Y. Acad. Sci. 1989, 570, 514–517. [Google Scholar] [CrossRef]
- Ha, S.-B. Phytochelatin Synthase Genes from Arabidopsis and the Yeast Schizosaccharomyces pombe. Plant Cell 1999, 11, 1153–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, S.B.; Kodliwadmath, M.V.; Kodliwadmath, S.M. Study of oxidative stress and enzymatic antioxidants in normal pregnancy. Ind. J. Clin. Biochem. 2007, 22, 135–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, D.; Meng, H.; Ge, J.; Zhang, L.; Wang, H.; Bai, D.; Li, J.; Qu, L.; Li, Z. Ultrasensitive fluorometric glutathione assay based on a conformational switch of a G-quadruplex mediated by silver(I). Microchim. Acta 2017, 184, 3325–3332. [Google Scholar] [CrossRef]
- Xiangzhao, A.; Qiang, M.; Xingguang, S. Nanosensor for dopamine and glutathione based on the quenching and recovery of the fluorescence of silica-coated quantum dots. Microchim. Acta 2012, 180, 269–277. [Google Scholar] [CrossRef]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Aspects Med. 2009, 30, 42–59. [Google Scholar] [CrossRef] [Green Version]
- Staal, F.; Ela, S.; Roederer, M.; Anderson, M.; Herzenberg, L. Glutathione deficiency and human immunodeficiency virus infection. Lancet 1992, 339, 909–912. [Google Scholar] [CrossRef]
- Samiec, P.S.; Drews-Botsch, C.; Flagg, E.W.; Kurtz, J.C.; Sternberg, P.; Reed, R.L.; Jones, D.P. Glutathione in Human Plasma: Decline in Association with Aging, Age-Related Macular Degeneration, and Diabetes. Free Radic. Biol. Med. 1998, 24, 699–704. [Google Scholar] [CrossRef]
- Tang, Z.; Chen, H.; He, H.; Ma, C. Tapiero, Assay for alkaline phosphatase activity: Progress and prospects. Trends Anal. Chem. 2019, 113, 32–43. [Google Scholar] [CrossRef]
- Park, K.S.; Kim, M.I.; Woo, M.A.; Park, H.G. A label-free method for detecting biological thiols based on blocking of Hg2+-quenching of fluorescent gold nanoclusters. Biosens. Bioelectron. 2013, 45, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Yuan, J.; Wang, E. Sensitive and Selective Sensor for Biothiols in the Cell Based on the Recovered Fluorescence of the CdTe Quantum Dots−Hg(II) System. Anal. Chem. 2009, 81, 5569–5573. [Google Scholar] [CrossRef] [PubMed]
- Heafield, M.T.; Fearn, S.; Steventon, G.B.; Waring, R.H.; Williams, A.C.; Sturman, S.G. Plasma cysteine and sulphate levels in patients with motor neurone, Parkinson’s and Alzheimer’s disease. Neurosci. Lett. 1990, 110, 216–220. [Google Scholar] [CrossRef]
- Cao, X.N.; Li, J.H.; Xu, H.H.; Lin, L.; Xian, Y.Z.; Yamamoto, K.; Jin, L.T. Platinum particles-modified electrode for HPLC with pulsed amperometric detection of thiols in rat striatum. Biomed. Chromatogr. 2010, 18, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.Y.; Liu, Z.Y.; Qi, L.M.; Lai, J.P.; Kitte, S.A.; Xu, G.B. Ultrasensitive Glutathione Detection Based on Lucigenin Cathodic Electrochemiluminescence in the Presence of MnO2 Nanosheets. Anal. Chim. 2016, 88, 7654–7659. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Chang, H.T. Nile Red-Adsorbed Gold Nanoparticle Matrixes for Determining Aminothiols through Surface-Assisted Laser Desorption/Ionization Mass Spectrometry. Anal. Chim. 2006, 78, 1485–1493. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Jana, N.R. Detection of Cellular Glutathione and Oxidized Glutathione Using Magnetic–Plasmonic Nanocomposite-Based “Turn-Off” Surface Enhanced Raman Scattering. Anal. Chem. 2013, 85, 9221–9228. [Google Scholar] [CrossRef] [PubMed]
- Khusbu, F.Y.; Zhou, X.; Chen, H.; Ma, C.; Wang, K. Thioflavin T as a fluorescence probe for biosensing applications. TrAC Trends Anal. Chem. 2018, 109, 1–18. [Google Scholar] [CrossRef]
- Ma, C.; Wu, K.; Zhao, H.; Liu, H.; Wang, K.; Xia, K. Fluorometric aptamer-based determination of ochratoxin A based on the use of graphene oxide and RNase H-aided amplification. Microchim. Acta 2018, 185, 347. [Google Scholar] [CrossRef]
- Chen, M.; Li, W.; Ma, C.; Wu, K.; He, H.; Wang, K. Fluorometric determination of the activity of uracil-DNA glycosylase by using graphene oxide and exonuclease I assisted signal amplification. Microchim. Acta 2019, 186, 110. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Ma, C.; Zhao, H.; Chen, M.; Deng, Z. Sensitive aptamer-based fluorescene assay for ochratoxin A based on RNase H signal amplification. Food Chem. 2019, 277, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Hepel, M. “Molecular Beacon”-Based Fluorescent Assay for Selective Detection of Glutathione and Cysteine. Anal. Chem. 2011, 83, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yao, T.; Shi, S.; Cao, Y.; Sun, W. A label-free fluorescent probe for Hg2+ and biothiols based on graphene oxide and Ru-complex. Sci. Rep. 2014, 4, 5320. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Huang, J.; Yang, X.; Yang, Y.; Quan, K.; Xie, N.; Li, J.; Ma, C.; Wang, K. Gold Nanoparticle Loaded Split-DNAzyme Probe for Amplified miRNA Detection in Living Cells. Anal. Chem. 2017, 89, 8377–8383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, Q.Y.; Li, J.; Ge, J.; Wang, J.Y.; Dong, Z.Z.; Li, Z.H. A label-free method for detecting biothiols based on poly(thymine)-templated copper nanoparticles. Biosens. Bioelectron. 2015, 69, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.H.; Ding, J.S.; Liu, J.W. 2-Aminopurine-modified DNA homopolymers for robust and sensitive detection of mercury and silver. Biosens. Bioelectron. 2017, 87, 171–177. [Google Scholar] [CrossRef]
- Zhao, H.; Xiang, X.; Chen, M.; Ma, C. Aptamer-Based Fluorometric Ochratoxin A Assay Based on Photoinduced Electron Transfer. Toxins 2019, 11, 65. [Google Scholar] [CrossRef]
- Wu, K.; Ma, C.; Deng, Z.; Fang, N.; Tang, Z.; Zhu, X.; Wang, K. Label-free and nicking enzyme-assisted fluorescence signal amplification for RNase H determination based on a G-quadruplexe/thioflavin T complex. Talanta 2018, 182, 142–147. [Google Scholar] [CrossRef]
- Wu, K.; Ma, C.; Zhao, H.; He, H.; Chen, H. Label-Free G-Quadruplex Aptamer Fluorescence Assay for Ochratoxin A Using a Thioflavin T Probe. Toxins 2018, 10, 198. [Google Scholar] [CrossRef]
- Ono, A.; Cao, S.; Togashi, H.; Tashiro, M.; Fujimoto, T.; Machinami, T.; Oda, S.; Miyake, Y.; Okamoto, I.; Tanaka, Y. Specific interactions between silver(i) ions and cytosine–cytosine pairs in DNA duplexes. Chem. Commun. 2008, 39, 4825. [Google Scholar] [CrossRef] [PubMed]
- Shamsipur, M.; Safavi, A.; Mohammadpour, Z. Indirect colorimetric detection of glutathione based on its radical restoration ability using carbon nanodots as nanozymes. Sens. Actuators B Chem. 2014, 199, 463–469. [Google Scholar] [CrossRef]
- Zhou, X.H.; Kong, D.M.; Shen, H.X. Ag+ and Cysteine Quantitation Based on G-Quadruplex-Hemin DNAzymes Disruption by Ag+. Anal. Chem. 2009, 82, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Wei, W.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. Conformational switch of G-quadruplex as a label-free platform for fluorescence detection of Ag+ and biothiol. Anal. Methods 2015, 8, 311–315. [Google Scholar] [CrossRef]
- Tong, L.L.; Li, L.; Chen, Z.Z.; Wang, Q.; Tang, B. Stable label-free fluorescent sensing of biothiols based on ThT direct inducing conformation-specific G-quadruplex. Biosens. Bioelectron. 2013, 49, 420–425. [Google Scholar] [CrossRef]





| Method | Material | Analyte | Linear Range (nM) | LOD (nM) | Reference |
|---|---|---|---|---|---|
| ECL | MnO2 Nanosheet | GSH | 10–2000 | 3.7 | [16] |
| Colorimetry | Carbon nanodot | GSH | 0–7000 | 300 | [32] |
| Colorimetry | DNAzyme | GSH | 40–1500 | 40 | [33] |
| Cys | 50–650 | 50 | |||
| Fluorescence | G-quadruplex | GSH | 50–3600 | 16 | [34] |
| Fluorescence | Thioflavin T | GSH | 30–2000 | 13.9 | [35] |
| Cys | 20–2500 | 8.4 | |||
| Fluorescence | Graphene oxide and Ru-complex | GSH | 0–5000 | 4.6 | [24] |
| Cys | 0–5000 | 6.2 | |||
| Fluorescence | 2-Aminopurine | GSH | 3–1000 | 3 | This work |
| Cys | 5–1000 | 5 |
| Sample | Added (μM) | Found (μM) | Recovery (%) |
|---|---|---|---|
| 1 | 0 | 0.29 ± 0.0034 | - |
| 2 | 0.4 | 0.68 ± 0.0006 | 97.5 |
| 3 | 0.5 | 0.80 ± 0.0031 | 102 |
| 4 | 0.6 | 0.88 ± 0.0017 | 98.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Chen, M.; Ma, C. Fluorescent Method for the Detection of Biothiols Using an Ag+-Mediated Conformational Switch. Sensors 2019, 19, 934. https://doi.org/10.3390/s19040934
Zhao H, Chen M, Ma C. Fluorescent Method for the Detection of Biothiols Using an Ag+-Mediated Conformational Switch. Sensors. 2019; 19(4):934. https://doi.org/10.3390/s19040934
Chicago/Turabian StyleZhao, Han, Mingjian Chen, and Changbei Ma. 2019. "Fluorescent Method for the Detection of Biothiols Using an Ag+-Mediated Conformational Switch" Sensors 19, no. 4: 934. https://doi.org/10.3390/s19040934




