Ion Chromatographic Fingerprinting of STC-1 Cellular Response for Taste Sensing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Preparation
2.2. Ion Chromatographic (IC) Analysis
2.3. STC-1 Cells’ Viability Evaluation
2.4. Data Analysis
3. Results and Discussion
3.1. Chromatograms of STC-1 Cellular Response Towards Taste Stimuli
3.2. Viability Study
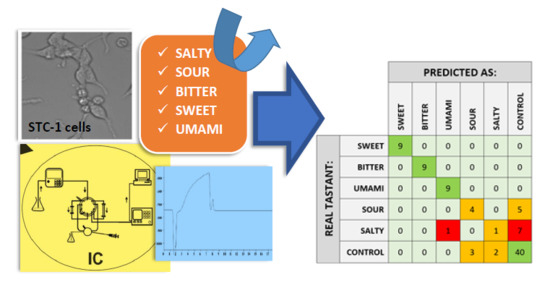
3.3. Taste Recognition Using PLS-DA
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chaudhari, N.; Roper, S.D. The cell biology of taste. J. Cell Biol. 2010, 190, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Suddendorf, T.; Bulley, A.; Miloyan, B. Prospection and natural selection. Curr. Opin. Behav. Sci. 2018, 24, 26–31. [Google Scholar] [CrossRef]
- Wu, C.; Du, L.; Zou, L.; Zhao, L.; Huang, L.; Wang, P. Recent advances in taste cell- and receptor-based biosensors. Sens. Actuators B Chem. 2014, 201, 75–85. [Google Scholar] [CrossRef]
- Ciosek, P.; Wróblewski, W. Sensor arrays for liquid sensing—Electronic tongue systems. Analyst 2007, 132, 963. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Hu, X.; Zhu, Z. Biomimetic sensors and biosensors for qualitative and quantitative analyses of five basic tastes. TrAC Trends Anal. Chem. 2017, 87, 58–70. [Google Scholar] [CrossRef]
- Chen, P.; Liu, X.; Wang, B.; Cheng, G.; Wang, P. A biomimetic taste receptor cell-based biosensor for electrophysiology recording and acidic sensation. Sens. Actuators B Chem. 2009, 139, 576–583. [Google Scholar] [CrossRef]
- Lee, J.S.; Cho, A.-N.; Jin, Y.; Kim, J.; Kim, S.; Cho, S.-W. Bio-artificial tongue with tongue extracellular matrix and primary taste cells. Biomaterials 2018, 151, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hui, G.; Deng, S. A novel sweet taste cell-based sensor. Biosens. Bioelectron. 2010, 26, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Son, M.; Park, T.H. The bioelectronic nose and tongue using olfactory and taste receptors: Analytical tools for food quality and safety assessment. Biotechnol. Adv. 2018, 36, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Lillehoj, P.B.; Wang, P. Bioanalytical and chemical sensors using living taste, olfactory, and neural cells and tissues: A short review. Analyst 2015, 140, 7048–7061. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Du, Y.-W.; Huang, L.; Ben-Shoshan Galeczki, Y.; Dagan-Wiener, A.; Naim, M.; Niv, M.; Wang, P. Biomimetic Sensors for the Senses: Towards Better Understanding of Taste and Odor Sensation. Sensors 2017, 17, 2881. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, Y.; Liu, Q.; Xu, Y.; Cai, H.; Wang, P. A novel experimental research based on taste cell chips for taste transduction mechanism. Sens. Actuators B Chem. 2008, 131, 24–28. [Google Scholar] [CrossRef]
- Hui, G.-H.; Mi, S.-S.; Deng, S.-P. Sweet and bitter tastants specific detection by the taste cell-based sensor. Biosens. Bioelectron. 2012, 35, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zou, L.; Zhao, L.; Wang, P.; Wu, C. Biomimetic chemical sensors using bioengineered olfactory and taste cells. Bioengineered 2014, 5, 326–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, L.; Zou, L.; Zhao, L.; Huang, L.; Wang, P.; Wu, C. Label-free functional assays of chemical receptors using a bioengineered cell-based biosensor with localized extracellular acidification measurement. Biosens. Bioelectron. 2014, 54, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Stone, R.C.; Stojadinovic, O.; Rosa, A.M.; Ramirez, H.A.; Badiavas, E.; Blumenberg, M.; Tomic-Canic, M. A bioengineered living cell construct activates an acute wound healing response in venous leg ulcers. Sci. Transl. Med. 2017, 9, eaaf8611. [Google Scholar] [CrossRef] [PubMed]
- Barham, H.P.; Cooper, S.E.; Anderson, C.B.; Tizzano, M.; Kingdom, T.T.; Finger, T.E.; Kinnamon, S.C.; Ramakrishnan, V.R. Solitary chemosensory cells and bitter taste receptor signaling in human sinonasal mucosa. Int. Forum Allergy Rhinol. 2013, 3, 450–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, G.; Mi, S.; Ye, S.; Jin, J.; Chen, Q.; Yu, Z. Tastant quantitative analysis from complex mixtures using taste cell-based sensor and double-layered cascaded series stochastic resonance. Electrochim. Acta 2014, 136, 75–88. [Google Scholar] [CrossRef]
- Chen, M.C.; Wu, S.V.; Reeve, J.R.; Rozengurt, E. Bitter stimuli induce Ca2+ signaling and CCK release in enteroendocrine STC-1 cells: Role of L-type voltage-sensitive Ca2+ channels. Am. J. Physiol. Physiol. 2006, 291, C726–C739. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.R.; An, J.H.; Jang, I.H.; Na, W.; Yang, H.; Cho, K.H.; Lee, S.H.; Song, H.S.; Jang, J.; Park, T.H. High-performance bioelectronic tongue using ligand binding domain T1R1 VFT for umami taste detection. Biosens. Bioelectron. 2018, 117, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Song, H.S.; Jin, H.J.; Ahn, S.R.; Kim, D.; Lee, S.H.; Kim, U.-K.; Simons, C.T.; Hong, S.; Park, T.H. Bioelectronic Tongue Using Heterodimeric Human Taste Receptor for the Discrimination of Sweeteners with Human-like Performance. ACS Nano 2014, 8, 9781–9789. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.R.; An, J.H.; Song, H.S.; Park, J.W.; Lee, S.H.; Kim, J.H.; Jang, J.; Park, T.H. Duplex Bioelectronic Tongue for Sensing Umami and Sweet Tastes Based on Human Taste Receptor Nanovesicles. ACS Nano 2016, 10, 7287–7296. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Du, L.; Zou, L.; Huang, L.; Wang, P. A biomimetic bitter receptor-based biosensor with high efficiency immobilization and purification using self-assembled aptamers. Analyst 2013, 138, 5989–5994. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Liang, J.; Gu, F.; Du, D.; Chen, F. Berberine activates bitter taste responses of enteroendocrine STC-1 cells. Mol. Cell. Biochem. 2018, 447, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Du, L.; Qin, Z.; Gao, K.; Wang, P. Dual extracellular recording using a light-addressable potentiometric sensor for taste signal transduction. In Proceedings of the 2017 ISOCS/IEEE International Symposium on Olfaction and Electronic Nose (ISOEN), Montreal, QC, Canada, 28–31 May 2017. [Google Scholar]
- Pihlström, T.; Valverde, A.; Reynolds, S.; Fernández-Alba, A.R.; Mol, H.; Chemist, S.; Jezussek Bavarian Health, M.; Stajnbaher, D.; Valenciana, G.; Paula Medina, S.; et al. Method Validation and Quality Control Procedures for Pesticide Residues Analysis in Food and Feed. SANCO, Document N SANCO/12495/2011. Supersedes Document No. SANCO/10684/2009 Implemented by 01/01/2012. Available online: http://www.crl-pesticides.eu/library/docs/fv/SANCO12495-2011.pdf (accessed on 1 March 2019).
- Wiriyawattana, P.; Suwonsichon, S.; Suwonsichon, T. Effects of aging on taste thresholds: A case of Asian people. J. Sens. Stud. 2018, 33, e12436. [Google Scholar] [CrossRef]
- Weiß, J. Handbook of Ion Chromatography, 4th ed.; Wiley: Weinheim, Germany, 2016; ISBN 9783527651641. [Google Scholar]
- Stockert, J.C.; Blázquez-Castro, A.; Cañete, M.; Horobin, R.W.; Villanueva, Á. MTT assay for cell viability: Intracellular localization of the formazan product is in lipid droplets. Acta Histochem. 2012, 114, 785–796. [Google Scholar] [CrossRef] [PubMed]
- ISO 10993-5:2009 Biological Evaluation of medical devices—Part 5: Tests for in vitro Cytotoxicity. Available online: https://www.iso.org/standard/36406.html (accessed on 31 December 2018).
- Zabadaj, M.; Ufnalska, I.; Chreptowicz, K.; Mierzejewska, J.; Wróblewski, W.; Ciosek-Skibińska, P. Performance of hybrid electronic tongue and HPLC coupled with chemometric analysis for the monitoring of yeast biotransformation. Chemom. Intell. Lab. Syst. 2017, 167, 69–77. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zabadaj, M.; Szuplewska, A.; Balcerzak, M.; Chudy, M.; Ciosek-Skibińska, P. Ion Chromatographic Fingerprinting of STC-1 Cellular Response for Taste Sensing. Sensors 2019, 19, 1062. https://doi.org/10.3390/s19051062
Zabadaj M, Szuplewska A, Balcerzak M, Chudy M, Ciosek-Skibińska P. Ion Chromatographic Fingerprinting of STC-1 Cellular Response for Taste Sensing. Sensors. 2019; 19(5):1062. https://doi.org/10.3390/s19051062
Chicago/Turabian StyleZabadaj, Marcin, Aleksandra Szuplewska, Maria Balcerzak, Michał Chudy, and Patrycja Ciosek-Skibińska. 2019. "Ion Chromatographic Fingerprinting of STC-1 Cellular Response for Taste Sensing" Sensors 19, no. 5: 1062. https://doi.org/10.3390/s19051062
APA StyleZabadaj, M., Szuplewska, A., Balcerzak, M., Chudy, M., & Ciosek-Skibińska, P. (2019). Ion Chromatographic Fingerprinting of STC-1 Cellular Response for Taste Sensing. Sensors, 19(5), 1062. https://doi.org/10.3390/s19051062