Deep Learning-Based Detection of Pigment Signs for Analysis and Diagnosis of Retinitis Pigmentosa
Abstract
:1. Introduction
- -
- RPS-Net avoids the postprocessing step to enhance segmentation results.
- -
- RPS-Net utilizes deep-feature concatenation inside and outside of the encoder-decoder to enhance the quality of the feature.
- -
- For fair comparison with other research results, the trained RPS-Net models and algorithms are made publicly available through [26].
2. Related Works
2.1. Retinal Image Segmentation Based on Handcrafted Features
2.2. Retinal Image Segmentation Based on Deep-Feature (CNN)
3. Proposed Method
3.1. Overview of the Proposed Architecture
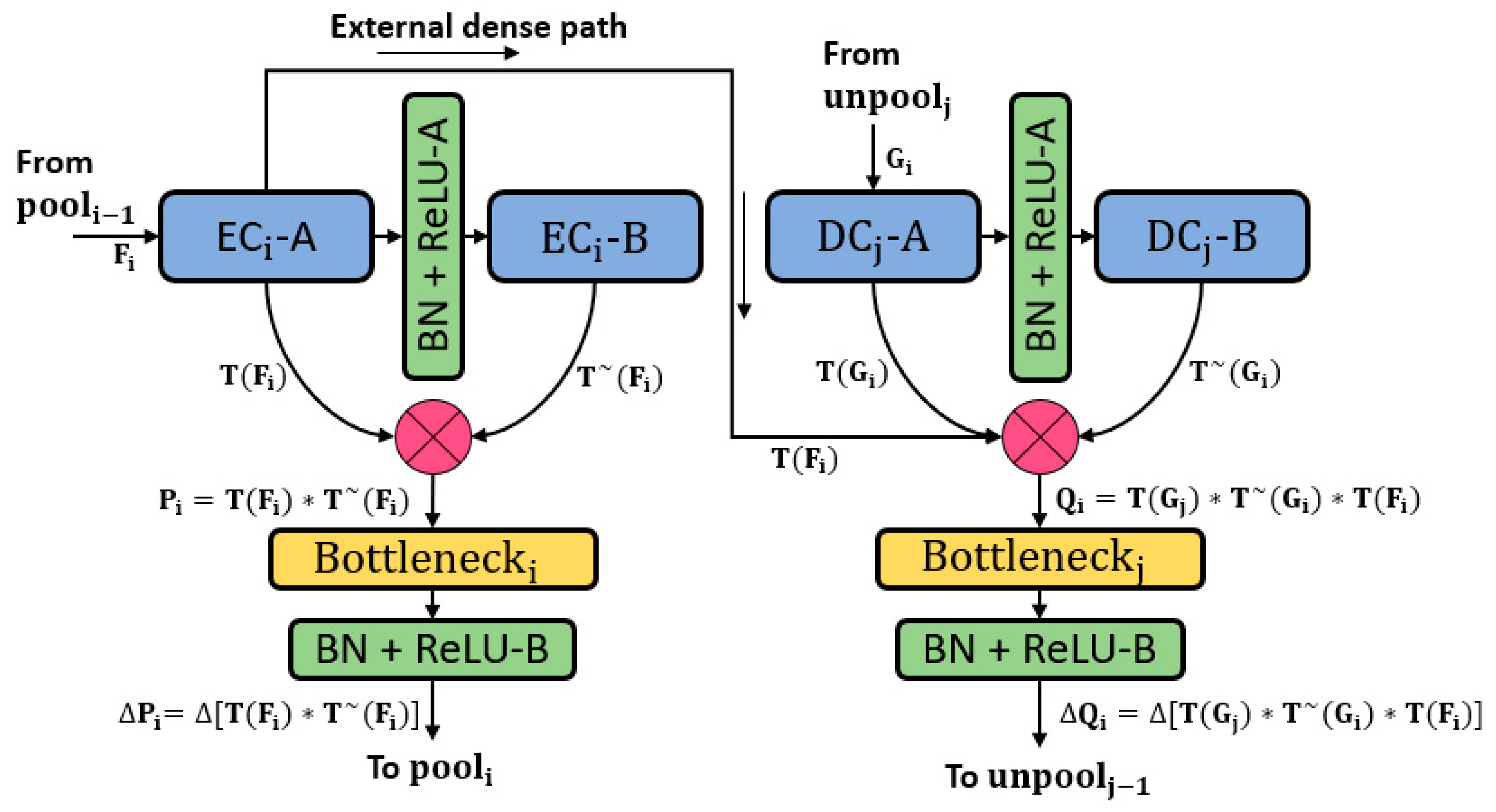
3.2. Retinal Pigment Sign Segmentation Using RPS-Net
4. Experimental Results
4.1. Experimental Data and Environment
4.2. Data Augmentation
4.3. RPS-Net Training
4.4. Testing of the Proposed Method
4.4.1. RPS-Net Testing for Pigment Sign Segmentation
4.4.2. Retinal Pigment Sign Segmentation Results by RPS-Net
4.4.3. Comparison of RPS-Net with Other Methods
5. Discussion
5.1. Detection/Counting and Size Analysis of Retinal Pigments
5.2. Location Analysis for PS
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Badar, M.; Haris, M.; Fatima, A. Application of deep learning for retinal image analysis: A review. Comput. Sci. Rev. 2020, 35, 100203. [Google Scholar] [CrossRef]
- Narayan, D.S.; Wood, J.P.M.; Chidlow, G.; Casson, R.J. A review of the mechanisms of cone degeneration in retinitis pigmentosa. Acta Ophthalmol. 2016, 94, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Schuerch, K.; Marsiglia, M.; Lee, W.; Tsang, S.H.; Sparrow, J.R. Multimodal imaging of disease-associated pigmentary changes in retinitis pigmentosa. Retina 2016, 36, S147–S158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limoli, P.G.; Vingolo, E.M.; Limoli, C.; Nebbioso, M. Stem cell surgery and growth factors in retinitis pigmentosa patients: Pilot study after literature review. Biomedicines 2019, 7, 94. [Google Scholar] [CrossRef] [Green Version]
- Menghini, M.; Cehajic-Kapetanovic, J.; MacLaren, R.E. Monitoring progression of retinitis pigmentosa: Current recommendations and recent advances. Expert Opin. Orphan Drugs 2020, 8, 67–78. [Google Scholar] [CrossRef]
- Son, J.; Shin, J.Y.; Kim, H.D.; Jung, K.-H.; Park, K.H.; Park, S.J. Development and validation of deep learning models for screening multiple abnormal findings in retinal fundus images. Ophthalmology 2020, 127, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Das, A.; Giri, R.; Chourasia, G.; Bala, A.A. Classification of retinal diseases using transfer learning approach. In Proceedings of the International Conference on Communication and Electronics Systems, Coimbatore, India, 17–19 July 2019; pp. 2080–2084. [Google Scholar]
- Bhatkalkar, B.J.; Reddy, D.R.; Prabhu, S.; Bhandary, S.V. Improving the performance of convolutional neural network for the segmentation of optic disc in fundus images using attention gates and conditional random fields. IEEE Access 2020, 8, 29299–29310. [Google Scholar] [CrossRef]
- Maji, D.; Santara, A.; Ghosh, S.; Sheet, D.; Mitra, P. Deep neural network and random forest hybrid architecture for learning to detect retinal vessels in fundus images. In Proceedings of the 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Milan, Italy, 25–29 August 2015; pp. 3029–3032. [Google Scholar]
- Xiuqin, P.; Zhang, Q.; Zhang, H.; Li, S. A fundus retinal vessels segmentation scheme based on the improved deep learning U-Net model. IEEE Access 2019, 7, 122634–122643. [Google Scholar] [CrossRef]
- Wang, L.; Liu, H.; Lu, Y.; Chen, H.; Zhang, J.; Pu, J. A coarse-to-fine deep learning framework for optic disc segmentation in fundus images. Biomed. Signal Process. Control 2019, 51, 82–89. [Google Scholar] [CrossRef]
- Kim, J.; Tran, L.; Chew, E.Y.; Antani, S. Optic disc and cup segmentation for glaucoma characterization using deep learning. In Proceedings of the IEEE 32nd International Symposium on Computer-Based Medical Systems, Cordoba, Spain, 5–7 June 2019; pp. 489–494. [Google Scholar]
- Edupuganti, V.G.; Chawla, A.; Kale, A. Automatic optic disk and cup segmentation of fundus images using deep learning. In Proceedings of the 25th IEEE International Conference on Image Processing, Athens, Greece, 7–10 October 2018; pp. 2227–2231. [Google Scholar]
- Rehman, Z.U.; Naqvi, S.S.; Khan, T.M.; Arsalan, M.; Khan, M.A.; Khalil, M.A. Multi-parametric optic disc segmentation using superpixel based feature classification. Expert Syst. Appl. 2019, 120, 461–473. [Google Scholar] [CrossRef]
- Arsalan, M.; Owais, M.; Mahmood, T.; Cho, S.W.; Park, K.R. Aiding the diagnosis of diabetic and hypertensive retinopathy using artificial intelligence-based semantic segmentation. J. Clin. Med. 2019, 8, 1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iftikhar, M.; Lemus, M.; Usmani, B.; Campochiaro, P.A.; Sahel, J.A.; Scholl, H.P.N.; Shah, S.M.A. Classification of disease severity in retinitis pigmentosa. Br. J. Ophthalmol. 2019, 103, 1595–1599. [Google Scholar] [CrossRef]
- Wintergerst, M.W.M.; Petrak, M.; Li, J.Q.; Larsen, P.P.; Berger, M.; Holz, F.G.; Finger, R.P.; Krohne, T.U. Non-contact smartphone-based fundus imaging compared to conventional fundus imaging: A low-cost alternative for retinopathy of prematurity screening and documentation. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaidehi, K.; Srilatha, J. A review on automatic glaucoma detection in retinal fundus images. In Proceedings of the 3rd International Conference on Data Engineering and Communication Technology, Hyderabad, India, 15–16 March 2019; pp. 485–498. [Google Scholar]
- Yan, Z.; Han, X.; Wang, C.; Qiu, Y.; Xiong, Z.; Cui, S. Learning mutually local-global U-Nets for high-resolution retinal lesion segmentation in fundus images. In Proceedings of the IEEE 16th International Symposium on Biomedical Imaging, Venice, Italy, 8–11 April 2019; pp. 597–600. [Google Scholar]
- Alheejawi, S.; Mandal, M.; Xu, H.; Lu, C.; Berendt, R.; Jha, N. Deep Learning Techniques for Biomedical and Health Informatics; Agarwal, B., Balas, V.E., Jain, L.C., Poonia, R.C., Manisha, Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 237–265. [Google Scholar]
- Amin, J.; Sharif, A.; Gul, N.; Anjum, M.A.; Nisar, M.W.; Azam, F.; Bukhari, S.A.C. Integrated design of deep features fusion for localization and classification of skin cancer. Pattern Recognit. Lett. 2020, 131, 63–70. [Google Scholar] [CrossRef]
- Owais, M.; Arsalan, M.; Choi, J.; Mahmood, T.; Park, K.R. Artificial intelligence-based classification of multiple gastrointestinal diseases using endoscopy videos for clinical diagnosis. J. Clin. Med. 2019, 8, 986. [Google Scholar] [CrossRef] [Green Version]
- Owais, M.; Arsalan, M.; Choi, J.; Park, K.R. Effective diagnosis and treatment through content-based medical image retrieval (CBMIR) by using artificial intelligence. J. Clin. Med. 2019, 8, 462. [Google Scholar] [CrossRef] [Green Version]
- Mahmood, T.; Arsalan, M.; Owais, M.; Lee, M.B.; Park, K.R. Artificial intelligence-based mitosis detection in breast cancer histopathology images using faster R-CNN and deep CNNs. J. Clin. Med. 2020, 9, 749. [Google Scholar] [CrossRef] [Green Version]
- Arsalan, M.; Owais, M.; Mahmood, T.; Choi, J.; Park, K.R. Artificial intelligence-based diagnosis of cardiac and related diseases. J. Clin. Med. 2020, 9, 871. [Google Scholar] [CrossRef] [Green Version]
- RPS-Net Model with Algoritm. Available online: http://dm.dgu.edu/link.html (accessed on 30 March 2020).
- Sánchez, C.I.; Hornero, R.; López, M.I.; Aboy, M.; Poza, J.; Abásolo, D. A novel automatic image processing algorithm for detection of hard exudates based on retinal image analysis. Med. Eng. Phys. 2008, 30, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Thibault, G.; Decencière, E.; Marcotegui, B.; Laÿ, B.; Danno, R.; Cazuguel, G.; Quellec, G.; Lamard, M.; Massin, P.; et al. Exudate detection in color retinal images for mass screening of diabetic retinopathy. Med. Image Anal. 2014, 18, 1026–1043. [Google Scholar] [CrossRef] [Green Version]
- Welfer, D.; Scharcanski, J.; Marinho, D.R. A coarse-to-fine strategy for automatically detecting exudates in color eye fundus images. Comput. Med. Imaging Graph. 2010, 34, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Götzinger, E.; Pircher, M.; Geitzenauer, W.; Ahlers, C.; Baumann, B.; Michels, S.; Schmidt-Erfurth, U.; Hitzenberger, C.K. Retinal pigment epithelium segmentation by polarization sensitive optical coherence tomography. Opt. Express 2008, 16, 16410–16422. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Reisman, C.A.; Chan, K.; Ramachandran, R.; Raza, A.; Hood, D.C. Automated segmentation of outer retinal layers in macular OCT images of patients with retinitis pigmentosa. Biomed. Opt. Express 2011, 2, 2493–2503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, H.; Saha, A.; Deb, S. An expert system to distinguish a defective eye from a normal eye. In Proceedings of the International Conference on Issues and Challenges in Intelligent Computing Techniques, Ghaziabad, India, 7–8 February 2014; pp. 155–158. [Google Scholar]
- Ravichandran, G.; Nath, M.K.; Elangovan, P. Diagnosis of retinitis pigmentosa from retinal images. Int. J. Electron. Telecommun. 2019, 65, 519–525. [Google Scholar]
- Guo, S.; Wang, K.; Kang, H.; Liu, T.; Gao, Y.; Li, T. Bin loss for hard exudates segmentation in fundus images. Neurocomputing 2019. In Press. [Google Scholar] [CrossRef]
- Mo, J.; Zhang, L.; Feng, Y. Exudate-based diabetic macular edema recognition in retinal images using cascaded deep residual networks. Neurocomputing 2018, 290, 161–171. [Google Scholar] [CrossRef]
- Prentašić, P.; Lončarić, S. Detection of exudates in fundus photographs using deep neural networks and anatomical landmark detection fusion. Comput. Methods Programs Biomed. 2016, 137, 281–292. [Google Scholar] [CrossRef]
- Tan, J.H.; Fujita, H.; Sivaprasad, S.; Bhandary, S.V.; Rao, A.K.; Chua, K.C.; Acharya, U.R. Automated segmentation of exudates, haemorrhages, microaneurysms using single convolutional neural network. Inf. Sci. 2017, 420, 66–76. [Google Scholar] [CrossRef]
- Sengupta, S.; Singh, A.; Leopold, H.A.; Gulati, T.; Lakshminarayanan, V. Ophthalmic diagnosis using deep learning with fundus images—A critical review. Artif. Intell. Med. 2020, 102, 101758. [Google Scholar] [CrossRef]
- Chudzik, P.; Majumdar, S.; Calivá, F.; Al-Diri, B.; Hunter, A. Microaneurysm detection using fully convolutional neural networks. Comput. Methods Programs Biomed. 2018, 158, 185–192. [Google Scholar] [CrossRef]
- Phasuk, S.; Poopresert, P.; Yaemsuk, A.; Suvannachart, P.; Itthipanichpong, R.; Chansangpetch, S.; Manassakorn, A.; Tantisevi, V.; Rojanapongpun, P.; Tantibundhit, C. Automated glaucoma screening from retinal fundus image using deep learning. In Proceedings of the 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Berlin, Germany, 23–27 July 2019; pp. 904–907. [Google Scholar]
- Christopher, M.; Bowd, C.; Belghith, A.; Goldbaum, M.H.; Weinreb, R.N.; Fazio, M.A.; Girkin, C.A.; Liebmann, J.M.; Zangwill, L.M. Deep learning approaches predict glaucomatous visual field damage from OCT optic nerve head en face images and retinal nerve fiber layer thickness maps. Ophthalmology 2020, 127, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.; Cardoso, J.S.; Soares, F. Offline computer-aided diagnosis for Glaucoma detection using fundus images targeted at mobile devices. Comput. Methods Programs Biomed. 2020, 192, 105341. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Cheng, J.; Xu, Y.; Wong, D.W.K.; Liu, J.; Cao, X. Joint optic disc and cup segmentation based on multi-label deep network and polar transformation. IEEE Trans. Med. Imaging 2018, 37, 1597–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Yu, L.; Yang, X.; Fu, C.-W.; Heng, P.-A. Patch-based output space adversarial learning for joint optic disc and cup segmentation. IEEE Trans. Med. Imaging 2019, 38, 2485–2495. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.M.; Yang, H.-C.; Poly, T.N.; Jian, W.-S.; Li, Y.-C. Deep learning algorithms for detection of diabetic retinopathy in retinal fundus photographs: A systematic review and meta-analysis. Comput. Methods Programs Biomed. 2020, 191, 105320. [Google Scholar] [CrossRef]
- Brancati, N.; Frucci, M.; Gragnaniello, D.; Riccio, D.; Di Iorio, V.; Di Perna, L.; Simonelli, F. Learning-based approach to segment pigment signs in fundus images for retinitis pigmentosa analysis. Neurocomputing 2018, 308, 159–171. [Google Scholar] [CrossRef]
- Brancati, N.; Frucci, M.; Riccio, D.; Di Perna, L.; Simonelli, F. Segmentation of pigment signs in fundus images for retinitis pigmentosa analysis by using deep learning. In Proceedings of the Image Analysis and Processing, Trento, Italy, 9–13 September 2019; pp. 437–445. [Google Scholar]
- Park, B.; Jeong, J. Color filter array demosaicking using densely connected residual network. IEEE Access 2019, 7, 128076–128085. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Arsalan, M.; Kim, D.S.; Owais, M.; Park, K.R. OR-Skip-Net: Outer residual skip network for skin segmentation in non-ideal situations. Expert Syst. Appl. 2020, 141, 112922. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.-W.; Heng, P.-A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Granada, Spain, 20 September 2018; pp. 3–11. [Google Scholar]
- GeForce GTX TITAN X Graphics Processing Unit. Available online: https://www.geforce.com/hardware/desktop-gpus/geforce-gtx-titan-x/specifications (accessed on 30 March 2020).
- MATLAB 2019b. Available online: https://ch.mathworks.com/downloads/web_downloads/download_release?release=R2019b (accessed on 23 March 2020).
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the International Conference for Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]







| Type | Methods | Strength | Limitation |
|---|---|---|---|
| RP by handcrafted features | Das et al. [32] | Uses simple image processing schemes. | Preprocessing is required. |
| Ravichandran et al. [33] | Watershed transform gives better region approximation. | The handcrafted feature-based method performance is subject to preprocessing by CLACHE. | |
| RP by learned features | Brancati et al. [46] | The simple machine learning classifier are used, AdaBoost provides less false negatives | The classification accuracy of the classifier is based on the, denoising, shade correction, etc. |
| Brancati et al. (Modified U-Net) [47] | Subsequently improved segmentation performance by modified U-Net model, and 15% improvement in F-measure compared to [46]. | The method performance is affected by more false negatives (represented by sensitivity of the method) compared to [46]. | |
| RPS-Net(Proposed) | Utilizes deep concatenation inside encoder-decoder, and encoder-to-decoder (outer) for immediate feature transfer and enhancement, with substantial reduction in false negatives. | Training for fully convolutional network requires a large amount of data by augmentation. |
| Method | Other Architectures | RPS-Net |
|---|---|---|
| SegNet [51] | Collectively, the network has 26 convolutional layers. | Total 16 convolutional layers (3 × 3) are used in the encoder and decoder with concatenation in each dense block. |
| No feature reuse policy is employed. | Dense connectivity in both the encoder and decoder for feature empowerment. | |
| First two dense blocks have two convolutional layers, whereas the others have three convolutional layers. | Each dense block similarly has two convolutional layers. | |
| The convolutional layer with 512-depth is utilized twice in the network. | The convolutional layer with 512-depth is used once for each encoder and decoder. | |
| OR-Skip-Net [52] | No feature reused policy is implemented for internal convolutional blocks. | Internal dense connectivity for both encoder and decoder. |
| Only external residual skip paths are used. | Both internal and external dense paths are used by concatenation. | |
| No bottleneck layers are used. | Bottleneck layers are employed in each dense block. | |
| Total of four residual connections are used in total. | Overall, 20 dense connections are used internally and externally. | |
| Vess-Net [15] | Based on residual connectivity. | Based on dense connectivity. |
| Feature empowerment from the first convolutional layer is missed and has no internal or external residual connection. | Each layer is densely connected. | |
| No bottleneck layer is used. | Bottleneck layers are employed in each dense block. | |
| Collectively, 10 residual paths. | Overall, 20 dense connections are used internally and externally. | |
| U-Net [53] | Overall, 23 convolutional layers are employed. | Total 16 convolutional layers (3 × 3) are used in the encoder and decoder with concatenation in each dense block. |
| Up convolutions are for the expansion part to upsample the features. | Up convolutions are not used. | |
| Based on residual and dense connectivity. | Based on dense connectivity. | |
| Convolution with 1024-depth is used between the encoder and decoder. | 1024-depth convolutions are ignored to reduce the number of parameters. | |
| Cropping layer is employed for borders. | Cropping is not required; pooling indices keep the image size the same. | |
| Modified U-Net [47] | Overall, 3 blocks are used in each encoder and decoder | Overall, 4 blocks are used in each encoder and decoder |
| The up convolutions are used for upsampling | Unpooling layers are used to upsample | |
| The deep feature concatenation is just used encoder-to-decoder | Feature concatenation used inside both encoder/decoder and encoder-to-decoder | |
| The number of filters considered is 32 to 128 | The number of filters considered is 64 to 512 | |
| Dense-U-Net [54] | Total of 4 dense blocks are used inside encoder with 6, 12, 36, 24 convolutional layers in each block respectively | Total 16 convolutional layers for overall network with occurrence of two convolutional layers in each block |
| Average pooling used in each encoder block | Max pooling used in each encoder block | |
| Five up convolutions are used in decoder for upsampling | 4 unpooling layers are used in decoder for upsampling | |
| H-Dense-U-Net [54] | Combines 2-D Dene-U-Net and 3-D Dene-U-Net for voxel wise prediction | Used for pixel wise prediction |
| Total 4 dense blocks are used inside encoder with 3, 4, 12, 8 3-D convolutional layers in combination of 2-D Dense-U-net fusion | Total of 16 2-D convolutional layers for overall network | |
| Designed for 3-D volumetric features | Designed for 2-D image features | |
| Utilizes 3-D average pooling layer in each 3-D dense block | Used 2-Maxpooling layer in each encoder dense block | |
| U-Net++ [55] | The external dense path is with dense convolutional block | No convolutional layer is used in external dense path |
| There is a pyramid type structure of dense convolutional blocks between the encoder and decoder | Direct flat dense paths are used | |
| Individual dense blocks in dense path also have own dense skip connections | No convolutions are used in dense skip path |
| Block | Name/Size | Number of Filters | Output Feature Map Size (Width × Height × Number of Channels) |
|---|---|---|---|
| Encoder DB-1 | EC1-A ^^/3 × 3 × 3 To decoder (EDP-1) and Ecat-1 | 64 | 300 × 400 × 64 |
| EC1-B/3 × 3 × 64 To Ecat-1 | 64 | ||
| Ecat-1 (EC1-A * EC1-B) | - | 300 × 400 × 128 | |
| Bneck-1^^/1 × 1 × 64 | 300 × 400 × 64 | ||
| Pool-1 | - | 150 × 200 × 64 | |
| Encoder DB-1 | EC2-A ^^/3 × 3 × 64 To decoder (EDP-2) and Ecat-2 | 128 | 150 × 200 × 128 |
| EC2-B/3 × 3 × 64 To Ecat-2 | 128 | ||
| Ecat-2 (EC2-A * EC2-B) | - | 150 × 200 × 256 | |
| Bneck-2^^/1 × 1 × 128 | 150 × 200 × 128 | ||
| Pool-2 | - | 75 × 100 × 128 | |
| Encoder DB-1 | EC3-A ^^/3 × 3 × 64 To decoder (EDP-3) and Ecat-3 | 256 | 75 × 100 × 256 |
| EC3-B/3 × 3 × 64 To Ecat-3 | 256 | ||
| Ecat-3 (EC3-A * EC3-B) | - | 75 × 100 × 512 | |
| Bneck-3^^/1 × 1 × 256 | 75 × 100 × 256 | ||
| Pool-3 | - | 37 × 50 × 256 | |
| Encoder DB-1 | EC4-A ^^/3 × 3 × 64 To decoder (EDP-4) and Ecat-4 | 512 | 37 × 50 × 512 |
| EC4-B/3 × 3 × 64 To Ecat-4 | 512 | ||
| Ecat-4 (EC4-A * EC4-B) | - | 37 × 50 × 1024 | |
| Bneck-4^^/1 × 1 × 512 | 37 × 50 × 512 | ||
| Pool-4 | - | 18 × 25 × 512 |
| Block | Name/Size | Number of Filters | Output Feature Map Size (Width × Height × Number of Channels) |
|---|---|---|---|
| Decoder DB-4 | Unpool-4 | - | 37 × 50 × 512 |
| DC4-B ^^/3 × 3 × 512 To Dcat-4 | 512 | ||
| DC4-A/3 × 3 × 512 To Dcat-4 | 256 | 37 × 50 × 256 | |
| Dcat-4 (DC4-B * DC4-A * EC4-A) | - | 37 × 50 × 1280 | |
| Bneck-5^^/1 × 1 × 1280 | 256 | 37 × 50 × 256 | |
| Decoder DB-3 | Unpool-3 | - | 75 × 100 × 256 |
| DC3-B ^^/3 × 3 × 256 To Dcat-3 | 256 | ||
| DC3-A/3 × 3 × 256 To Dcat-3 | 128 | 75 × 100 × 128 | |
| Dcat-3 (DC3-B * DC3-A * EC3-A) | - | 75 × 100 × 640 | |
| Bneck-6^^/1 × 1 × 640 | 128 | 75 × 100 × 128 | |
| Decoder DB-2 | Unpool-2 | - | 150 × 200 × 128 |
| DC2-B ^^/3 × 3 × 128 To Dcat-2 | 128 | ||
| DC2-A/3 × 3 × 128 To Dcat-2 | 64 | 150 × 200 × 64 | |
| Dcat-2 (DC2-B * DC2-A * EC2-A) | - | 150 × 200 × 320 | |
| Bneck-7^^/1 × 1 × 320 | 64 | 150 × 200 × 64 | |
| Unpool-1 DB-1 | Unpool-1 | - | 300 × 400 × 64 |
| DC1-B ^^/3 × 3 × 64 To Dcat-1 | 64 | ||
| DC1-A/3 × 3 × 64 To Dcat-1 | 2 | 300 × 400 × 2 | |
| Dcat-1 (DC1-B * DC1-A * EC1-A) | - | 300 × 400 × 130 | |
| Bneck-8^^/1 × 1 × 130 | 2 | 300 × 400 × 2 | |
| Type | Method | Sen | Spe | P | F | Acc |
|---|---|---|---|---|---|---|
| Handcrafted local feature-based methods | * Ravichandran et al. [33] | 72.0 | 97.0 | - | 62.0 | 96.0 |
| Learned/deep-feature-based methods | Random Forest [46] | 58.26 | 99.46 | 46.18 | 47.93 | 99.14 |
| AdaBoost M1 [46] | 64.29 | 99.30 | 42.45 | 46.76 | 99.01 | |
| U-Net 48 × 48 [47] | 55.70 | 99.40 | 48.00 | 50.60 | 99.00 | |
| U-Net 72 × 72 [47] | 62.60 | 99.30 | 46.50 | 52.80 | 99.00 | |
| U-Net 96 × 96 [47] | 55.20 | 99.60 | 56.10 | 55.10 | 99.20 | |
| RPS-Net (proposed method) | 80.54 | 99.60 | 54.05 | 61.54 | 99.52 |
| Type | Method | Sen | Spe | P | F | Acc |
|---|---|---|---|---|---|---|
| Learned/deep-feature-based methods | Random Forest [46] | 56.20 | 99.48 | 50.49 | 49.29 | 99.11 |
| AdaBoost M1 [46] | 61.76 | 99.33 | 46.29 | 48.30 | 98.99 | |
| RPS-Net (proposed method) | 78.09 | 99.62 | 56.84 | 62.62 | 99.51 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arsalan, M.; Baek, N.R.; Owais, M.; Mahmood, T.; Park, K.R. Deep Learning-Based Detection of Pigment Signs for Analysis and Diagnosis of Retinitis Pigmentosa. Sensors 2020, 20, 3454. https://doi.org/10.3390/s20123454
Arsalan M, Baek NR, Owais M, Mahmood T, Park KR. Deep Learning-Based Detection of Pigment Signs for Analysis and Diagnosis of Retinitis Pigmentosa. Sensors. 2020; 20(12):3454. https://doi.org/10.3390/s20123454
Chicago/Turabian StyleArsalan, Muhammad, Na Rae Baek, Muhammad Owais, Tahir Mahmood, and Kang Ryoung Park. 2020. "Deep Learning-Based Detection of Pigment Signs for Analysis and Diagnosis of Retinitis Pigmentosa" Sensors 20, no. 12: 3454. https://doi.org/10.3390/s20123454
APA StyleArsalan, M., Baek, N. R., Owais, M., Mahmood, T., & Park, K. R. (2020). Deep Learning-Based Detection of Pigment Signs for Analysis and Diagnosis of Retinitis Pigmentosa. Sensors, 20(12), 3454. https://doi.org/10.3390/s20123454






