High-Throughput Phenotyping Approach for Screening Major Carotenoids of Tomato by Handheld Raman Spectroscopy Using Chemometric Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tomato Samples
2.2. Raman Spectral Data Acquisition
2.3. Profiling Carotenoids by HPLC
2.4. Quantification of Carotenoids by UV-Visible Spectrophotometry
2.5. Chemometric Analysis
2.5.1. Partial Least Square Regression (PLSR)
2.5.2. Soft Independent Modelling of Class Analogy (SIMCA)
2.5.3. Classification by Artificial Neural Networks (ANN)
3. Results
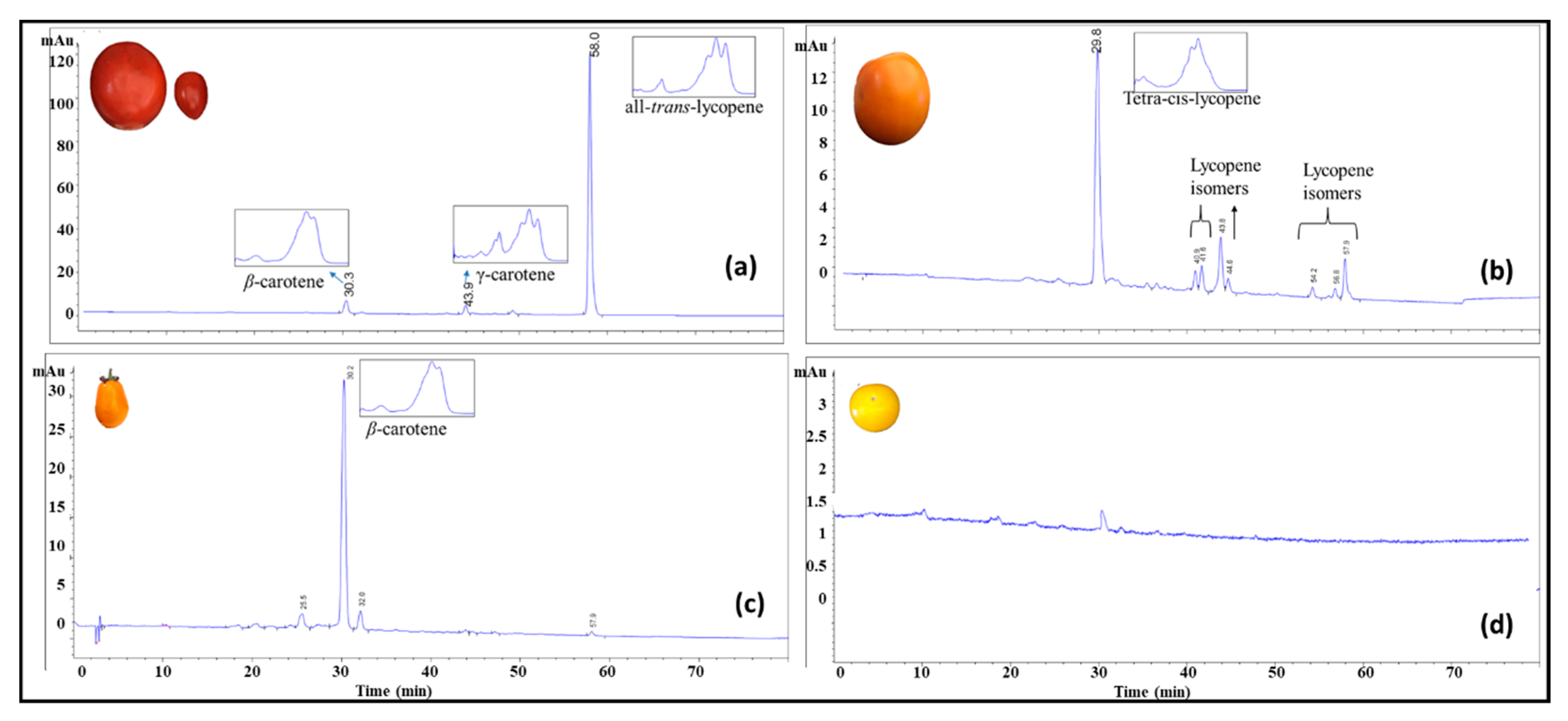
3.1. Characterization of the Carotenoid Profiles of Tomatoes
3.2. Quantification of All-Trans-Lycopene and β-Carotene by PLSR and ANN
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pek, Z.; Helyes, L.; Gyulai, G.; Foshee, W.; Daood, H.; Lau, J.; Waters, L.j. Molecular Profiling—Fruit Carotenoids Components of Six American Heirloom Tomatoes (Solanum lycopersicum). J. Forensic Biomech. 2016, 7. [Google Scholar] [CrossRef]
- Borguini, R.G.; da Silva Torres, E.A.F. Tomatoes and tomato products as dietary sources of antioxidants. Food Rev. Int. 2009, 25, 313–325. [Google Scholar] [CrossRef]
- Kopec, R.E.; Cooperstone, J.L.; Cichon, M.J.; Schwartz, S.J. Analysis Methods of Carotenoids. Anal. Antioxid. Rich Phytochem. 2012, 4, 105–148. [Google Scholar] [CrossRef]
- Luterotti, S.; Marković, K.; Franko, M.; Bicanic, D.; Madžgalj, A.; Kljak, K. Comparison of spectrophotometric and HPLC methods for determination of carotenoids in foods. Food Chem. 2013, 140, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Saltveit, M.E. Fruit Ripening and Fruit Quality. In Tomatoes; Heuvelink, E., Ed.; CABI Publishing: Wallingford, UK, 2005. [Google Scholar]
- Furbank, R.T.; Tester, M. Phenomics—technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Qin, J.; Chao, K.; Kim, M.S. Investigation of Raman chemical imaging for detection of lycopene changes in tomatoes during postharvest ripening. J. Food Eng. 2011, 107, 277–288. [Google Scholar] [CrossRef]
- Dzakovich, M.P.; Gas-Pascual, E.; Orchard, C.J.; Sari, E.N.; Riedl, K.M.; Schwartz, S.J.; Francis, D.M.; Cooperstone, J.L. Analysis of Tomato Carotenoids: Comparing Extraction and Chromatographic Methods. J. Aoac Int. 2019, 102, 1069–1079. [Google Scholar] [CrossRef]
- Fraser, P.D.; Enfissi, E.M.A.; Goodfellow, M.; Eguchi, T.; Bramley, P.M. Metabolite profiling of plant carotenoids using the matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Plant. J. 2007, 49, 552–564. [Google Scholar] [CrossRef]
- Matsubara, A.; Bamba, T.; Ishida, H.; Fukusaki, E.; Hirata, K. Highly sensitive and accurate profiling of carotenoids by supercritical fluid chromatography coupled with mass spectrometry. J. Sep. Sci. 2009, 32, 1459–1464. [Google Scholar] [CrossRef]
- Tiziani, S.; Schwartz, S.J.; Vodovotz, Y. Profiling of carotenoids in tomato juice by one- and two-dimensional NMR. J. Agric. Food Chem. 2006, 54, 6094–6100. [Google Scholar] [CrossRef]
- Schulz, H.; Baranska, M.; Baranski, R. Potential of NIR-FT-Raman spectroscopy in natural carotenoid analysis. Biopolymers 2005, 77, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Bhosale, P.; Ermakov, I.V.; Ermakova, M.R.; Gellermann, W.; Bernstein, P.S. Resonance Raman quantification of nutritionally important carotenoids in fruits, vegetables, and their juices in comparison to high-pressure liquid chromatography analysis. J. Agric. Food Chem. 2004, 52, 3281–3285. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Diaz, D.E.; De Nardo, T.; Santos, A.; de Jesus, S.; Francis, D.; Rodriguez-Saona, L.E. Profiling of nutritionally important carotenoids from genetically-diverse tomatoes by infrared spectroscopy. Food Chem. 2010, 120, 282–289. [Google Scholar] [CrossRef]
- Qin, J.; Chao, K.; Kim, M.S. Nondestructive evaluation of internal maturity of tomatoes using spatially offset Raman spectroscopy. Postharvest Biol. Technol. 2012, 71, 21–31. [Google Scholar] [CrossRef]
- Trebolazabala, J.; Maguregui, M.; Morillas, H.; de Diego, A.; Madariaga, J.M. Portable Raman spectroscopy for an in-situ monitoring the ripening of tomato (Solanum lycopersicum) fruits. Spectrochim. Acta Part A 2017, 180, 138–143. [Google Scholar] [CrossRef]
- Anthon, G.; Barrett, D.M. Standardization of a rapid spectrophotometric method for lycopene analysis. Acta Hortic. 2007, 758, 111–128. [Google Scholar] [CrossRef]
- Abdi, H. Partial least squares regression and projection on latent structure regression (PLS Regression). Wiley Interdiscip. Rev. 2010, 2, 97–106. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Kvalheim, O.M.; Karstang, T.V. SIMCA—Classification by Means of Disjoint Cross Validated Principal Components Models. Data Handl. Sci. Technol. 1992, 9, 209–248. [Google Scholar] [CrossRef]
- Duckworth, J. Mathematical Data Preprocessing. In Near-Infrared Spectroscopy in Agriculture; Al-Amoodi, L., Ed.; American Society of Agronomy, Inc.; Crop Science Society of America, Inc.; Soil Science Society of America, Inc.: Madison, WI, USA, 2004; pp. 115–132. [Google Scholar]
- Wold, S.; Sjöström, M. SIMCA: A Method for Analyzing Chemical Data in Terms of Similarity and Analogy; ACS Publications: Washington, DC, USA, 1977; pp. 243–282. [Google Scholar] [CrossRef]
- Olivieri, A.C. Mathematical Pre-processing. In Introduction to Multivariate Calibration; Springer: Cham, Switzerland, 2018; pp. 139–158. [Google Scholar] [CrossRef]
- Brereton, R.G. Multivariate Pattern Recognition in Chemometrics: Illustrated by Case Studies. Available online: https://www.sciencedirect.com/bookseries/data-handling-in-science-and-technology/vol/9 (accessed on 20 June 2020).
- Debska, B.; Guzowska-Świder, B. Application of artificial neural network in food classification. Anal. Chim. Acta 2011, 705, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, D.; Todeschini, R. Multivariate classification qualitative analysis. In Infrared Spectroscopy for Food Quality Analysis and Control; Elsevier Inc.: Amsterdam, The Netherlands, 2009; Available online: https://pdfs.semanticscholar.org/b6bd/4d48199287fefb932c8dc1fb9d69686bf082.pdf (accessed on 20 June 2020).
- Díaz, C.; Conde, J.E.; Estévez, D.; Pérez Olivero, S.J.; Pérez Trujillo, J.P. Application of multivariate analysis and artificial neural networks for the differentiation of red wines from the Canary Islands according to the island of origin. J. Agric. Food Chem. 2003, 51, 4303–4307. [Google Scholar] [CrossRef] [PubMed]
- Prevolnik, M.; Andronikov, D.; Žlender, B.; Font-i-Furnols, M.; Novič, M.; Škorjanc, D.; Čandek-Potokar, M. Classification of dry-cured hams according to the maturation time using near infrared spectra and artificial neural networks. Meat Sci. 2014, 96, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Tanajura da Silva, C.E.; Filardi, V.L.; Pepe, I.M.; Chaves, M.A.; Santos, C.M.S. Classification of food vegetable oils by fluorimetry and artificial neural networks. Food Control. 2015, 47, 86–91. [Google Scholar] [CrossRef] [Green Version]
- Silva, S.F.; Anjos, C.A.R.; Cavalcanti, R.N.; Celeghini, R.M.D.S. Evaluation of extra virgin olive oil stability by artificial neural network. Food Chem. 2015, 179, 35–43. [Google Scholar] [CrossRef]
- Binetti, G.; Del Coco, L.; Ragone, R.; Zelasco, S.; Perri, E.; Montemurro, C.; Schena, F.P. Cultivar classification of Apulian olive oils: Use of artificial neural networks for comparing NMR, NIR and merceological data. Food Chem. 2017, 219, 131–138. [Google Scholar] [CrossRef]
- Benvidi, A.; Abbasi, S.; Gharaghani, S.; Dehghan Tezerjani, M.; Masoum, S. Spectrophotometric determination of synthetic colorants using PSO–GA-ANN. Food Chem. 2017, 220, 377–384. [Google Scholar] [CrossRef]
- Gutiérrez-Reguera, F.; Jurado, J.M.; Montoya-Mayor, R.; Ternero-Rodríguez, M. Geographical classification of Spanish bottled mineral waters by means of iterative models based on linear discriminant analysis and artificial neural networks. Neural Comput. Appl. 2018, 29, 459–468. [Google Scholar] [CrossRef]
- Behkami, S.; Zain, S.M.; Gholami, M.; Khir, M.F.A. Classification of cow milk using artificial neural network developed from the spectral data of single- and three-detector spectrophotometers. Food Chem. 2019, 294, 309–315. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Ichinohe, Y.; Yamada, S.; Miyazaki, N.; Saito, S. Neural network-based preprocessing to estimate the parameters of the X-ray emission of a single-temperature thermal plasma. Mon. Not. R. Astron. Soc. 2018, 475, 4739–4744. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Huang, J.F.; Wang, F.M.; Tang, Y.L. Comparison between back propagation neural network and regression models for the estimation of pigment content in rice leaves and panicles using hyperspectral data. Int. J. Remote Sens. 2007, 28, 3457–3478. [Google Scholar] [CrossRef]
- De Oliveira, V.E.; Castro, H.V.; Edwards, H.G.; de Oliveira, L.F.C. Carotenes and carotenoids in natural biological samples: A Raman spectroscopic analysis. J. Raman Spectrosc. 2010, 41, 642–650. [Google Scholar] [CrossRef]
- Vijayalakshmi, K.; Jha, A.; Dasgupta, J. Ultrafast Triplet Generation and its Sensitization Drives Efficient Photoisomerization of Tetra-cis-lycopene to All-trans-lycopene. J. Phys. Chem. B 2015, 119, 8669–8678. [Google Scholar] [CrossRef] [PubMed]
- Llansola-Portoles, M.J.; Pascal, A.A.; Robert, B. Electronic and vibrational properties of carotenoids: From in vitro to in vivo. J. R. Soc. Interface 2017, 14, 20170504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegelman, C.H.; McShane, M.J.; Goetz, M.J.; Motamedi, M.; Yue, Q.L.; Coté, G.L. Theoretical Justification of Wavelength Selection in PLS Calibration: Development of a New Algorithm. Anal. Chem. 1998, 70, 35–44. [Google Scholar] [CrossRef]
- Ramesh, S.; Vydeki, D. Recognition and classification of paddy leaf diseases using Optimized Deep Neural network with Jaya algorithm. Inf. Process. Agric. 2019. [Google Scholar] [CrossRef]
- Brown, C.D.; Vega-Montoto, L.; Wentzell, P.D. Derivative preprocessing and optimal corrections for baseline drift in multivariate calibration. Appl. Spectrosc. 2000, 54, 1055–1068. [Google Scholar] [CrossRef]
- Hara, R.; Ishigaki, M.; Kitahama, Y.; Ozaki, Y.; Genkawa, T. Excitation wavelength selection for quantitative analysis of carotenoids in tomatoes using Raman spectroscopy. Food Chem. 2018, 258, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Sheng, R.; Cheng, W.; Li, H.; Ali, S.; Akomeah Agyekum, A.; Chen, Q. Model development for soluble solids and lycopene contents of cherry tomato at different temperatures using near-infrared spectroscopy. Postharvest Biol. Technol. 2019, 156, 110952. [Google Scholar] [CrossRef]


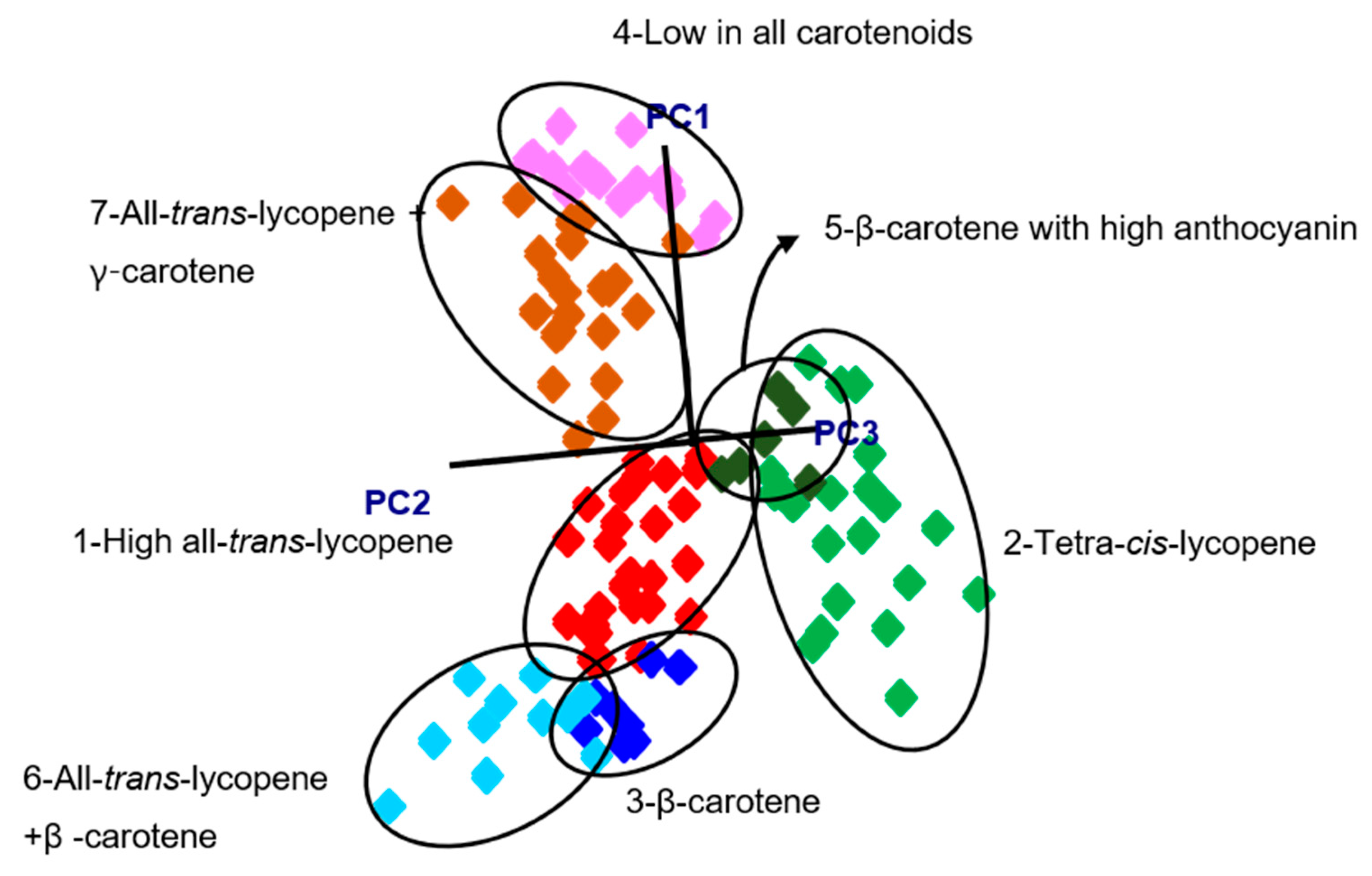

| All-Trans-Lycopene (High) | Tetra-Cis-Lycopene | β-Carotene | Low Carotenoids | β-Carotene + High Anthocyanin | All-Trans-Lycopene + β-Carotene | All-Trans-Lycopene (Low) | |
|---|---|---|---|---|---|---|---|
| all-trans-lycopene (high) | 0.0 | 3.4 | 6.1 | 18.4 | 5.7 | 7.5 | 6.2 |
| tetra-cis-lycopene | 0.0 | 6.9 | 8.7 | 4.7 | 10.7 | 7.1 | |
| β-carotene | 0.0 | 34.9 | 12.9 | 3.8 | 19.5 | ||
| low carotenoids | 0.0 | 9.9 | 38.5 | 5.4 | |||
| β-carotene with high anthocyanin | 0.0 | 17.0 | 6.3 | ||||
| all-trans-lycopene + β-carotene | 0.0 | 20.9 | |||||
| all-trans-lycopene (low) | 0.0 |
| Network Topology * | Dropout | Epoch | Classification Accuracy (%) | Test loss (CCE) |
|---|---|---|---|---|
| 60-242-7 | 0.4 | 814 | 100 | 0.03804 |
| 60-178-7 | 0.1 | 738 | 100 | 0.05484 |
| 60-240-7 | 1 | 746 | 100 | 0.06449 |
| 60-106-7 | 1 | 1417 | 100 | 0.06616 |
| 60-199-7 | 0.4 | 852 | 100 | 0.07257 |
| 60-226-7 | 0.1 | 499 | 100 | 0.07568 |
| 60-201-7 | 0.7 | 1388 | 100 | 0.07896 |
| 60-238-7 | 0.4 | 712 | 100 | 0.089558 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akpolat, H.; Barineau, M.; Jackson, K.A.; Akpolat, M.Z.; Francis, D.M.; Chen, Y.-J.; Rodriguez-Saona, L.E. High-Throughput Phenotyping Approach for Screening Major Carotenoids of Tomato by Handheld Raman Spectroscopy Using Chemometric Methods. Sensors 2020, 20, 3723. https://doi.org/10.3390/s20133723
Akpolat H, Barineau M, Jackson KA, Akpolat MZ, Francis DM, Chen Y-J, Rodriguez-Saona LE. High-Throughput Phenotyping Approach for Screening Major Carotenoids of Tomato by Handheld Raman Spectroscopy Using Chemometric Methods. Sensors. 2020; 20(13):3723. https://doi.org/10.3390/s20133723
Chicago/Turabian StyleAkpolat, Hacer, Mark Barineau, Keith A. Jackson, Mehmet Z. Akpolat, David M. Francis, Yu-Ju Chen, and Luis E. Rodriguez-Saona. 2020. "High-Throughput Phenotyping Approach for Screening Major Carotenoids of Tomato by Handheld Raman Spectroscopy Using Chemometric Methods" Sensors 20, no. 13: 3723. https://doi.org/10.3390/s20133723






