TumorGAN: A Multi-Modal Data Augmentation Framework for Brain Tumor Segmentation
Abstract
:1. Introduction
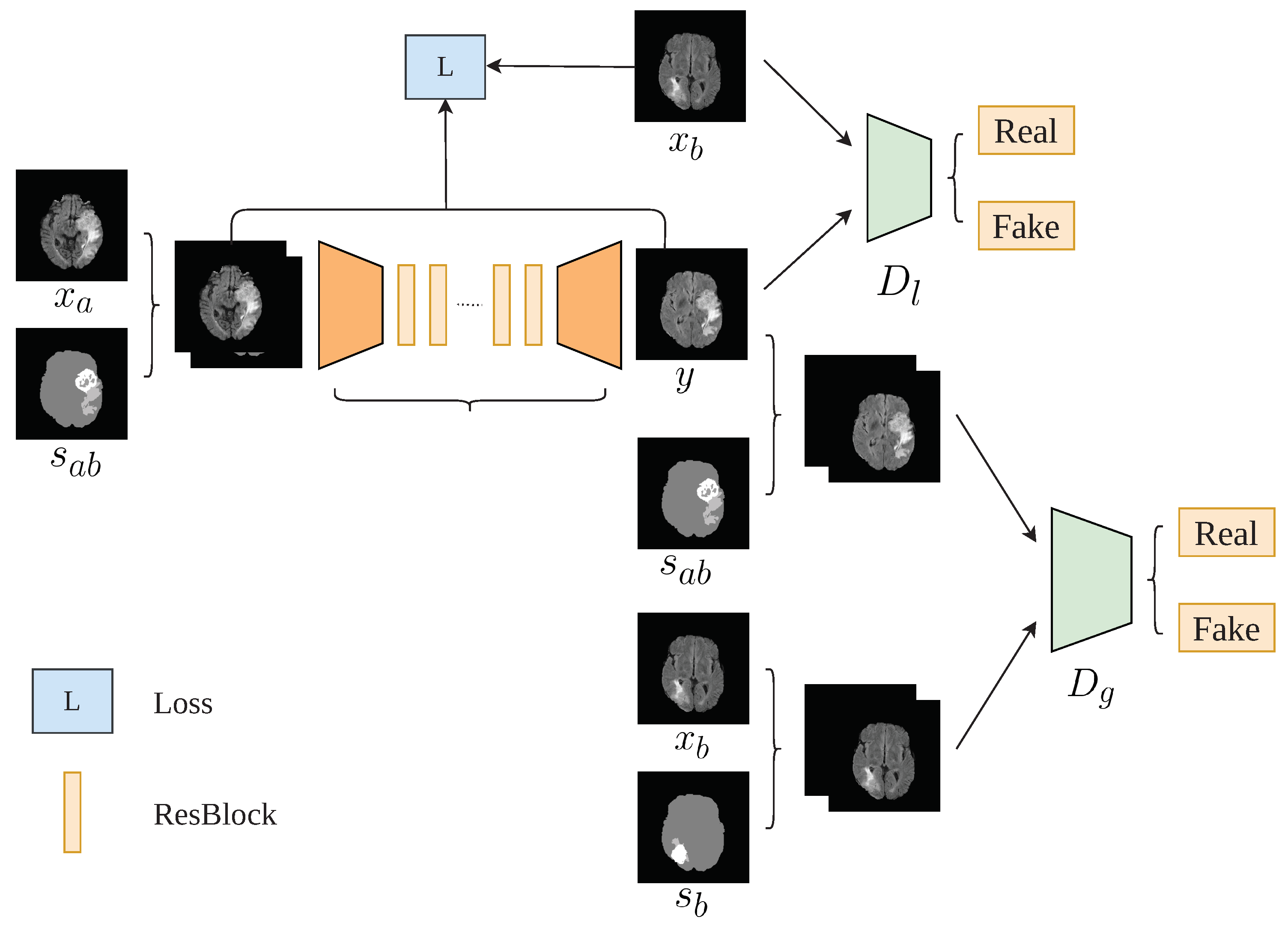
- We propose an image-to-image translation framework called TumorGAN, which can synthesize virtual image pairs from n real data pairs for brain tumor segmentation. Our experimental results show that TumorGAN is able to augment brain tumor data sets and can improve the performance of tumor segmentation for both single-modality data and multi-modal data.
- We design a region perceptual loss and an loss based on attention areas provided by the semantic labels to preserve the image details.
- We included an extra local discriminator co-operating with the main discriminator, in order to increase the efficiency of the discriminator and help TumorGAN to generate medical image pairs with more realistic details.
2. Related Work
2.1. Brain Tumor Segmentation
2.2. Generative Adversarial Network Based Medical Image Augmentation
3. Method
3.1. Synthesis of Semantic Label Image
3.2. Architecture
3.3. Formulation
4. Experiment
4.1. Implementation Details
4.2. Data Set Pre-Processing and Data Augmentation
4.3. Qualitative Evaluation
4.4. Tumor Segmentation Using Synthetic Data
4.4.1. Training on Multi-Modal Dataset
4.4.2. Training on Single Modality Data of U-Net
5. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
References
- Işın, A.; Direkoğlu, C.; Şah, M. Review of MRI-based brain tumor image segmentation using deep learning methods. Procedia Comput. Sci. 2016, 102, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Cordier, N.; Delingette, H.; Ayache, N. A patch-based approach for the segmentation of pathologies: Application to glioma labelling. IEEE Trans. Med. Imaging 2015, 35, 1066–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menze, B.H.; Van Leemput, K.; Lashkari, D.; Riklin-Raviv, T.; Geremia, E.; Alberts, E.; Gruber, P.; Wegener, S.; Weber, M.A.; Székely, G.; et al. A generative probabilistic model and discriminative extensions for brain lesion segmentation—with application to tumor and stroke. IEEE Trans. Med Imaging 2015, 35, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Ruan, S.; Canu, S. A review: Deep learning for medical image segmentation using multi-modality fusion. Array 2019, 3, 100004. [Google Scholar] [CrossRef]
- Zhou, C.; Ding, C.; Wang, X.; Lu, Z.; Tao, D. One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Trans. Image Process. 2020, 29, 4516–4529. [Google Scholar] [CrossRef] [Green Version]
- Tseng, K.L.; Lin, Y.L.; Hsu, W.; Huang, C.Y. Joint sequence learning and cross-modality convolution for 3d biomedical segmentation. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 22–25 July 2017; pp. 6393–6400. [Google Scholar]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef]
- Fu, M.; Wu, W.; Hong, X.; Liu, Q.; Jiang, J.; Ou, Y.; Zhao, Y.; Gong, X. Hierarchical combinatorial deep learning architecture for pancreas segmentation of medical computed tomography cancer images. BMC Syst. Biol. 2018, 12, 56. [Google Scholar] [CrossRef] [Green Version]
- Pfeiffer, M.; Funke, I.; Robu, M.R.; Bodenstedt, S.; Strenger, L.; Engelhardt, S.; Roß, T.; Clarkson, M.J.; Gurusamy, K.; Davidson, B.R.; et al. Generating large labeled data sets for laparoscopic image processing tasks using unpaired image-to-image translation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Advances in Neural Information Processing Systems; NIPS: Montreal, QC, Canada, 2014. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Chen, X.; Duan, Y.; Houthooft, R.; Schulman, J.; Sutskever, I.; Abbeel, P. Infogan: Interpretable representation learning by information maximizing generative adversarial nets. In Proceedings of the 30th International Conference on Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 2172–2180. [Google Scholar]
- Odena, A.; Olah, C.; Shlens, J. Conditional image synthesis with auxiliary classifier gans. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; Volume 70, pp. 2642–2651. [Google Scholar]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Sharma, M.M. Brain tumor segmentation techniques: A survey. Brain 2016, 4, 220–223. [Google Scholar]
- Rajendran, A.; Dhanasekaran, R. Fuzzy clustering and deformable model for tumor segmentation on MRI brain image: A combined approach. Procedia Eng. 2012, 30, 327–333. [Google Scholar] [CrossRef] [Green Version]
- Zabir, I.; Paul, S.; Rayhan, M.A.; Sarker, T.; Fattah, S.A.; Shahnaz, C. Automatic brain tumor detection and segmentation from multi-modal MRI images based on region growing and level set evolution. In Proceedings of the 2015 IEEE International WIE Conference on Electrical and Computer Engineering (WIECON-ECE), Dhaka, Bangladesh, 19–20 December 2015; pp. 503–506. [Google Scholar]
- Yousefi, S.; Azmi, R.; Zahedi, M. Brain tissue segmentation in MR images based on a hybrid of MRF and social algorithms. Med. Image Anal. 2012, 16, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Benson, C.; Deepa, V.; Lajish, V.; Rajamani, K. Brain tumor segmentation from MR brain images using improved fuzzy c-means clustering and watershed algorithm. In Proceedings of the 2016 International Conference on Advances in Computing, Communications and Informatics (ICACCI), Jaipur, India, 21–24 September 2016; pp. 187–192. [Google Scholar]
- Dvorak, P.; Menze, B. Structured prediction with convolutional neural networks for multimodal brain tumor segmentation. In Proceeding of the Multimodal Brain Tumor Image Segmentation Challenge, Munich, Germany, 5–9 October 2015; pp. 13–24. [Google Scholar]
- Colmeiro, R.R.; Verrastro, C.; Grosges, T. Multimodal brain tumor segmentation using 3D convolutional networks. In International MICCAI Brainlesion Workshop; Springer: Cham, Switzerland, 2017; pp. 226–240. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Shelhamer, E.; Long, J.; Darrell, T. Fully convolutional networks for semantic segmentation. IEEE Ann. Hist. Comput. 2017, 4, 640–651. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, UK, 2015. [Google Scholar]
- Cunniff, C.M.; Byrne, J.L.; Hudgins, L.M.; Moeschler, J.B.; Olney, A.H.; Pauli, R.M.; Seaver, L.H.; Stevens, C.A.; Figone, C. Informed consent for medical photographs. Genet. Med. Off. J. Am. Coll. Med. Genet. 2000, 2, 353–355. [Google Scholar]
- Simard, P.Y.; Steinkraus, D.; Platt, J.C. Best practices for convolutional neural networks applied to visual document analysis. Icdar 2003, 2, 958. [Google Scholar]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef] [Green Version]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Wolterink, J.M.; Dinkla, A.M.; Savenije, M.H.; Seevinck, P.R.; van den Berg, C.A.; Išgum, I. Deep MR to CT synthesis using unpaired data. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Quebec City, QC, Canada, 10–14 September 2017; pp. 14–23. [Google Scholar]
- Hiasa, Y.; Otake, Y.; Takao, M.; Matsuoka, T.; Takashima, K.; Carass, A.; Prince, J.L.; Sugano, N.; Sato, Y. Cross-modality image synthesis from unpaired data using CycleGAN. In Proceedings of the 32nd International Conference on Neural Information Processing Systems, Granada, Spain, 16–20 September 2018; pp. 31–41. [Google Scholar]
- Zhang, Z.; Yang, L.; Zheng, Y. Translating and segmenting multimodal medical volumes with cycle-and shape-consistency generative adversarial network. In Proceedings of the IEEE conference on computer vision and pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 9242–9251. [Google Scholar]
- Kang, E.; Koo, H.J.; Yang, D.H.; Seo, J.B.; Ye, J.C. Cycle-consistent adversarial denoising network for multiphase coronary CT angiography. Med. Phys. 2019, 46, 550–562. [Google Scholar] [CrossRef]
- Yi, X.; Babyn, P. Sharpness-aware low-dose CT denoising using conditional generative adversarial network. J. Digit. Imaging 2018, 31, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wang, C.; Yu, Z.; Zheng, H.; Zheng, B. Instance map based image synthesis with a denoising generative adversarial network. IEEE Access 2018, 6, 33654–33665. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, C.; Yu, Z.; Wang, N.; Zheng, H.; Zheng, B. Unpaired photo-to-caricature translation on faces in the wild. Neurocomputing 2019, 355, 71–81. [Google Scholar] [CrossRef] [Green Version]
- Ledig, C.; Theis, L.; Huszár, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-realistic single image super-resolution using a generative adversarial network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Mao, X.; Li, Q.; Xie, H.; Lau, R.Y.; Wang, Z.; Paul Smolley, S. Least squares generative adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision 2017, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In Proceedings of the International MICCAI Brainlesion Workshop, Quebec City, QC, Canada, 10–14 September 2017. [Google Scholar]
- Brügger, R.; Baumgartner, C.F.; Konukoglu, E. A Partially Reversible U-Net for Memory-Efficient Volumetric Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs trained by a two time-scale update rule converge to a local Nash equilibrium. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 6626–6637. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018. [Google Scholar]
- Fidon, L.; Li, W.; Garcia-Peraza-Herrera, L.C.; Ekanayake, J.; Kitchen, N.; Ourselin, S.; Vercauteren, T. Generalised wasserstein dice score for imbalanced multi-class segmentation using holistic convolutional networks. In Proceedings of the International MICCAI Brainlesion Workshop, Quebec City, QC, Canada, 10–14 September 2017; pp. 64–76. [Google Scholar]
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive growing of gans for improved quality, stability, and variation. arXiv 2017, arXiv:1710.10196. [Google Scholar]









| Data Sets | All | HGG | LGG |
|---|---|---|---|
| Total | 285 | 210 | 75 |
| Train | 226 | 166 | 60 |
| Augmentation | 226 | 166 | 60 |
| Test | 59 | 44 | 15 |
| CycleGAN (Baseline) | Pix2Pix | w/o per | w/o d_lcoal | TumorGAN | |
|---|---|---|---|---|---|
| FID | 154.86 (0%) | 126.42 (18.36%) | 87.75 (43.34%) | 145.67 (5.93%) | 77.43 (50%) |
| Networks | Whole | Core | en | Mean | |
|---|---|---|---|---|---|
| Cascaded Net | Without augmentation | 0.848 | 0.748 | 0.643 | 0.746 |
| With TumorGAN augmentation (ours) | 0.853 | 0.791 | 0.692 | 0.778 | |
| U-Net | Without augmentation | 0.783 | 0.672 | 0.609 | 0.687 |
| With TumorGAN augmentation (ours) | 0.806 | 0.704 | 0.611 | 0.706 | |
| Deeplab-v3 | Without augmentation | 0.820 | 0.700 | 0.571 | 0.697 |
| With TumorGAN augmentation (ours) | 0.831 | 0.762 | 0.584 | 0.725 | |
| Modality | Whole | Core | en | Mean | |
|---|---|---|---|---|---|
| flair | without augmentation | 0.754 | 0.513 | 0.286 | 0.518 |
| with pix2pix augmentation | 0.745 | 0.527 | 0.214 | 0.495 | |
| with TumorGAN augmentation | 0.765 | 0.522 | 0.289 | 0.525 | |
| t2 | without augmentation | 0.743 | 0.577 | 0.335 | 0.552 |
| with pix2pix augmentation | 0.729 | 0.593 | 0.220 | 0.514 | |
| with TumorGAN augmentation | 0.750 | 0.572 | 0.321 | 0.548 | |
| t1 | without augmentation | 0.628 | 0.422 | 0.199 | 0.416 |
| with pix2pix augmentation | 0.635 | 0.489 | 0.106 | 0.410 | |
| with TumorGAN augmentation | 0.628 | 0.467 | 0.235 | 0.443 | |
| t1ce | without augmentation | 0.597 | 0.534 | 0.570 | 0.567 |
| with pix2pix augmentation | 0.659 | 0.673 | 0.545 | 0.626 | |
| with TumorGAN augmentation | 0.671 | 0.681 | 0.589 | 0.647 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Yu, Z.; Wang, Y.; Zheng, H. TumorGAN: A Multi-Modal Data Augmentation Framework for Brain Tumor Segmentation. Sensors 2020, 20, 4203. https://doi.org/10.3390/s20154203
Li Q, Yu Z, Wang Y, Zheng H. TumorGAN: A Multi-Modal Data Augmentation Framework for Brain Tumor Segmentation. Sensors. 2020; 20(15):4203. https://doi.org/10.3390/s20154203
Chicago/Turabian StyleLi, Qingyun, Zhibin Yu, Yubo Wang, and Haiyong Zheng. 2020. "TumorGAN: A Multi-Modal Data Augmentation Framework for Brain Tumor Segmentation" Sensors 20, no. 15: 4203. https://doi.org/10.3390/s20154203
APA StyleLi, Q., Yu, Z., Wang, Y., & Zheng, H. (2020). TumorGAN: A Multi-Modal Data Augmentation Framework for Brain Tumor Segmentation. Sensors, 20(15), 4203. https://doi.org/10.3390/s20154203





