Sensing with Nanopores and Aptamers: A Way Forward
Abstract
:1. Introduction
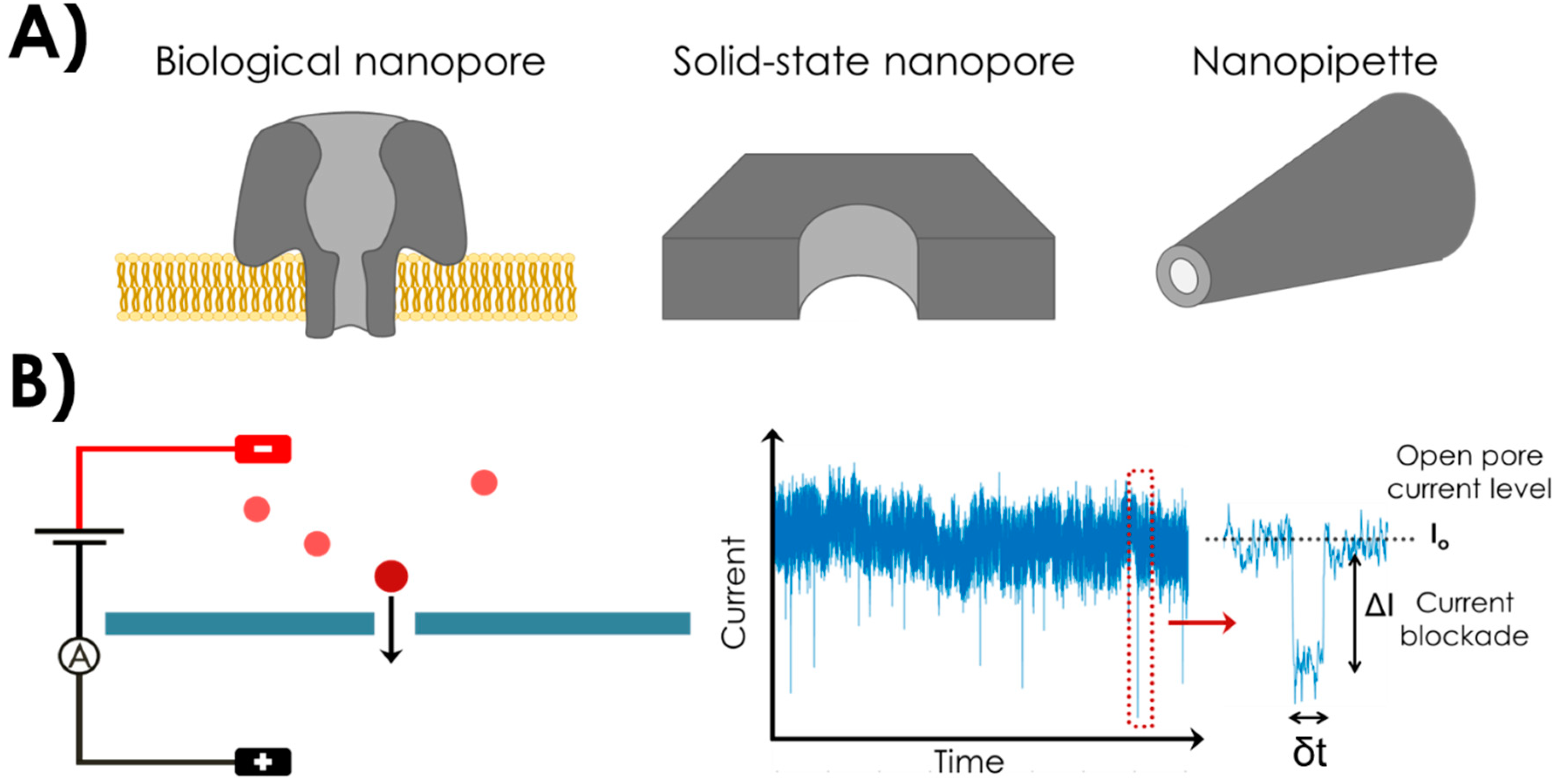
2. Nanopores and Nanopipettes
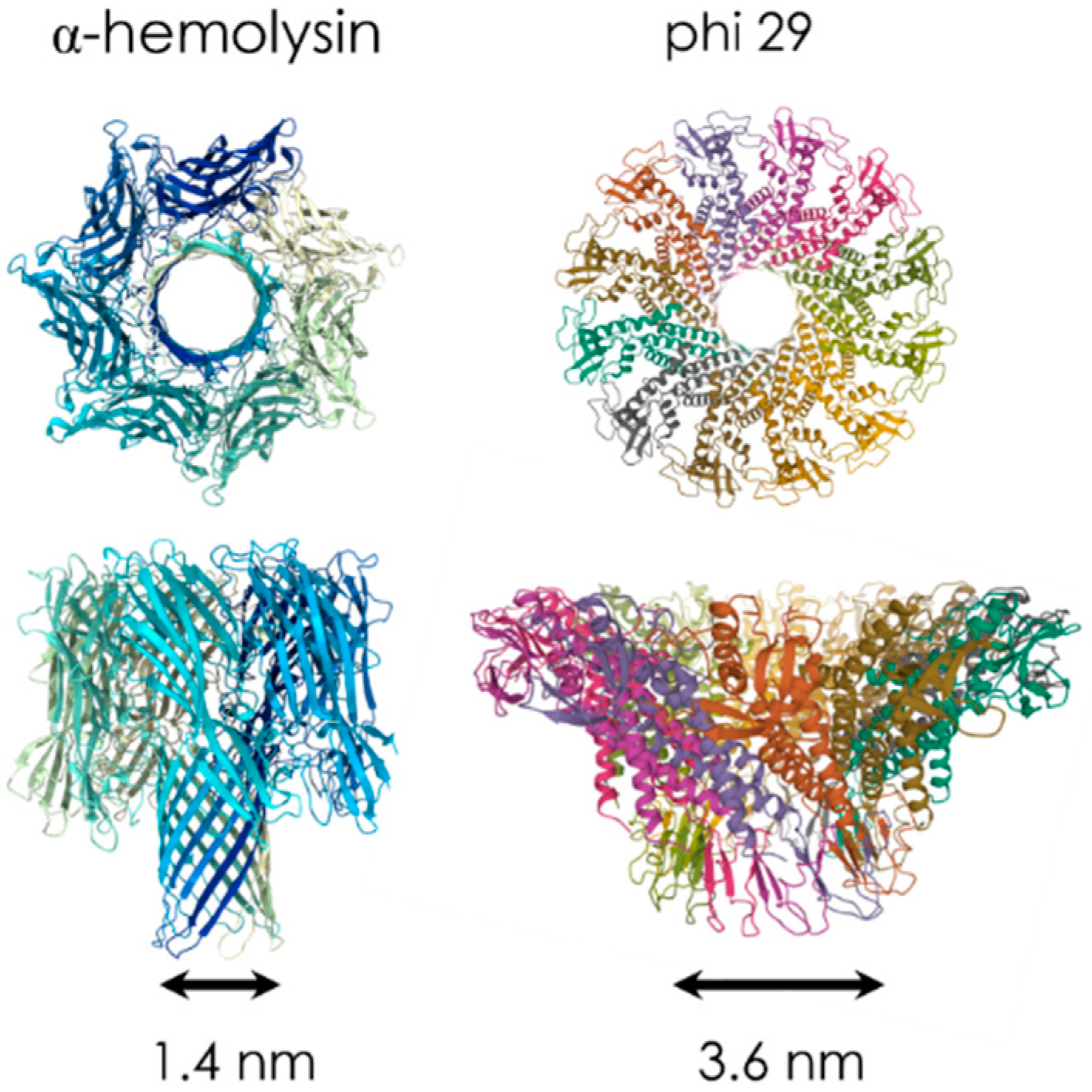
2.1. Biological Nanopores
2.2. Solid-State Nanopores
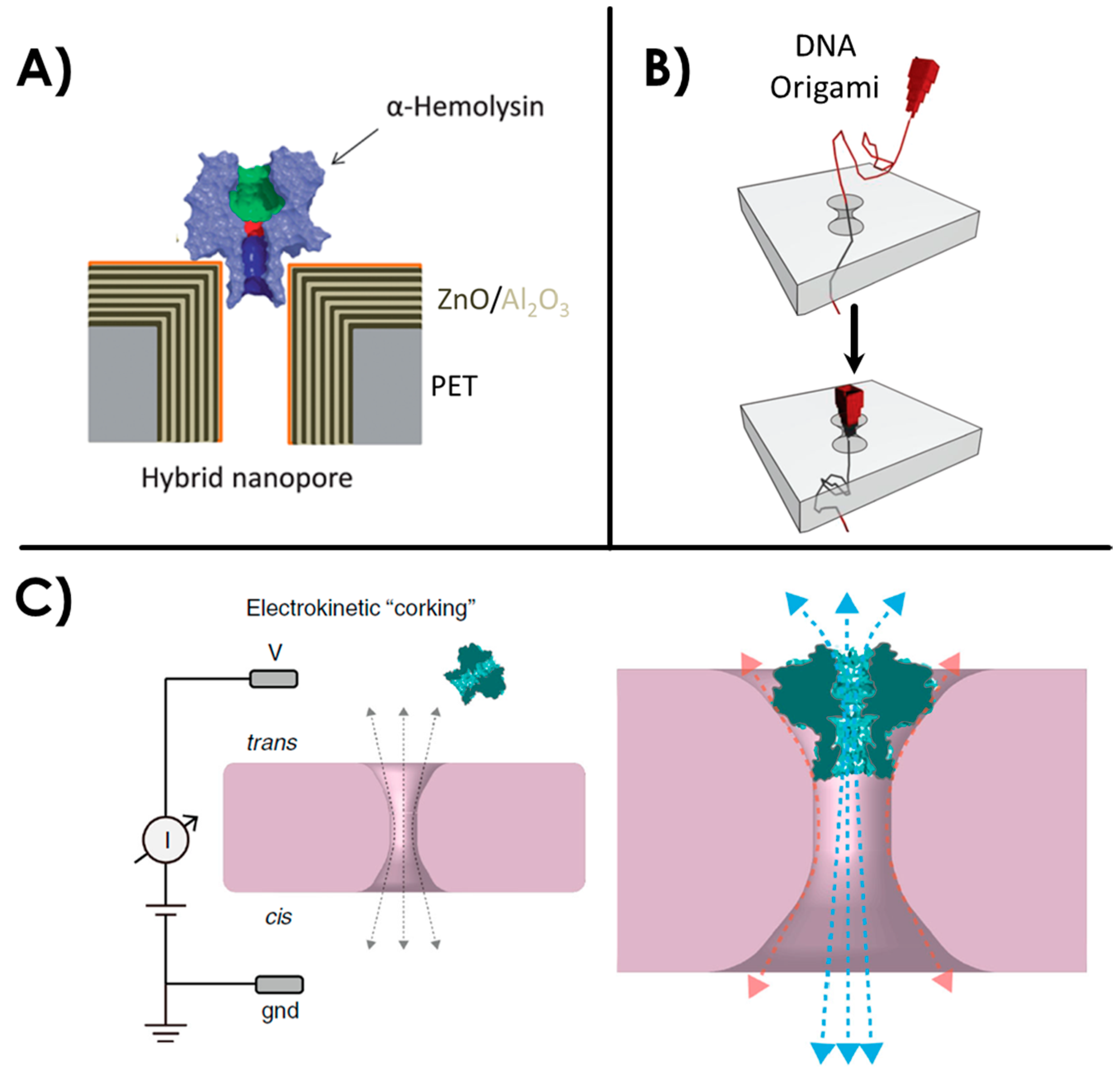
2.3. Hybrid or Biomimetic Nanopores
2.4. Nanopipettes or Glass Nanocapillaries
3. Nanopores and Aptamers: A Winning Combination
3.1. Aptamers: Molecular Swiss Army Knife
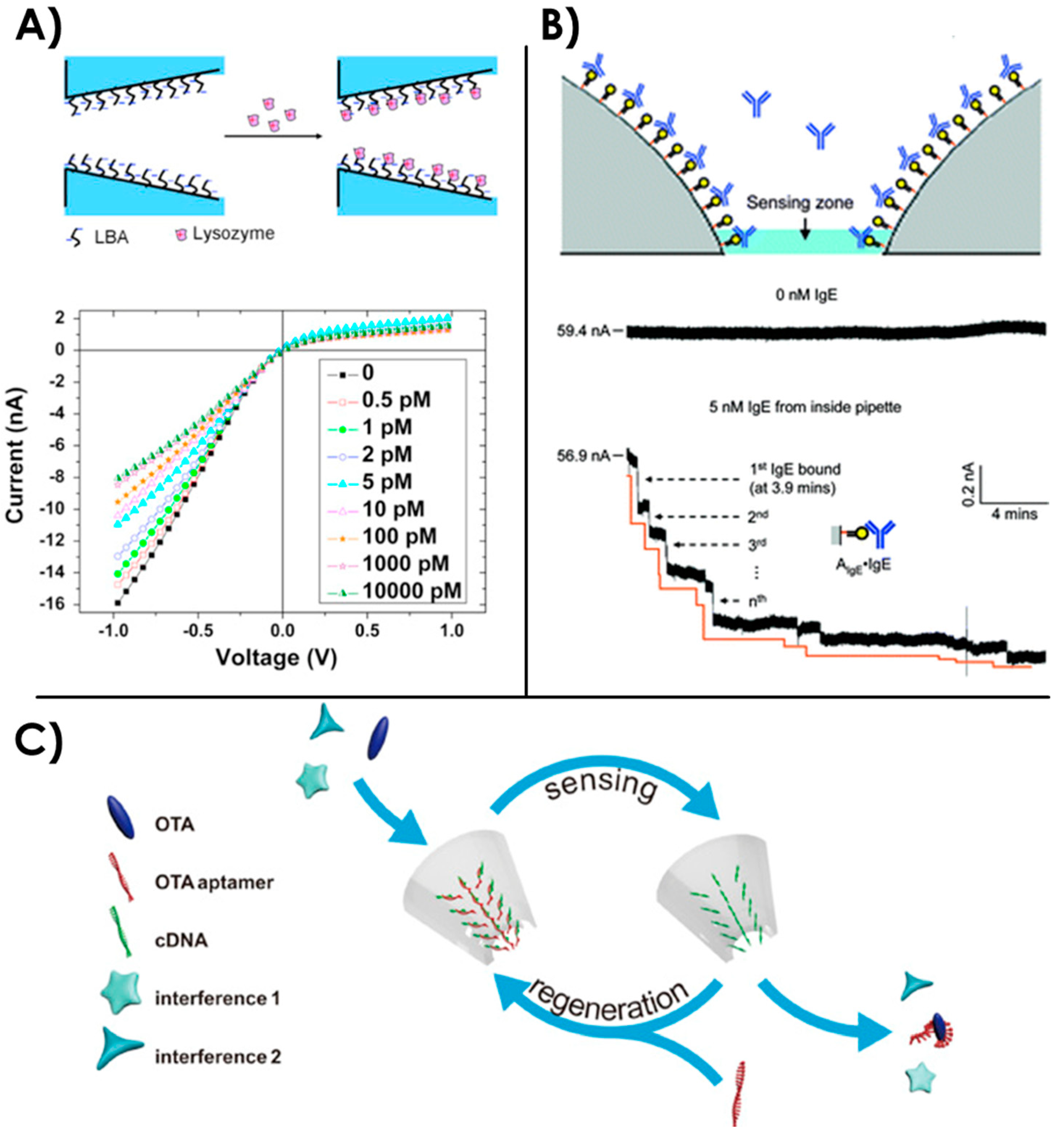
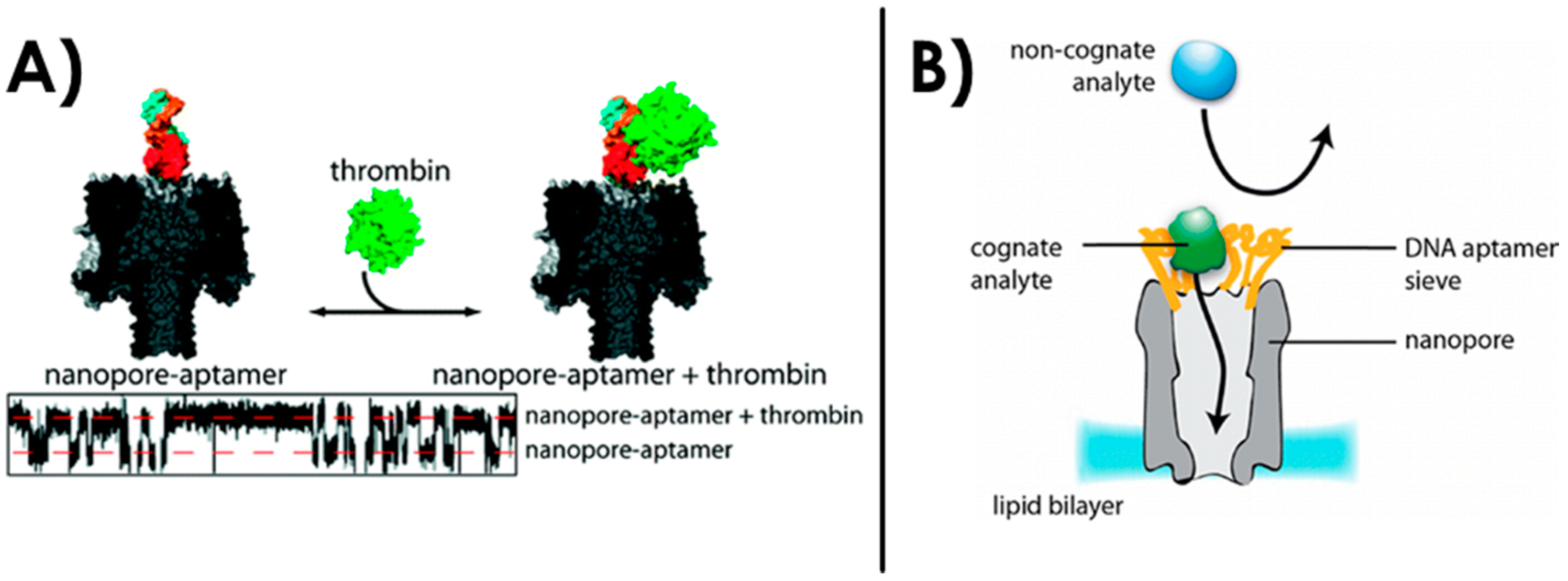
3.2. Aptamer Structure and Aptamer-Target Studies Using Naked Nanopores
3.3. Aptamers as Carrier Probes for Nanopore Sensing
3.3.1. Aptamers as Carriers for Nanopore Sensing of Their Target
3.3.2. Aptamer-Functionalized Nanoparticles as Carrier Probes for Nanopore Sensing
3.4. Sensing with Aptamer-Functionalized Nanopores
3.5. Gating with Aptamer-Functionalized Nanopores
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Coulter, W.H. Means for Counting Particles Suspended in a Fluid. U.S. Patent 26,565,08A, 20 October 1953. [Google Scholar]
- Hladky, S.B.; Haydon, D.A. Discreteness of Conductance Change in Bimolecular Lipid Membranes in the Presence of Certain Antibiotics. Nature 1970, 225, 451–453. [Google Scholar] [CrossRef] [PubMed]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Stein, D.; McMullan, C.; Branton, D.; Aziz, M.J.; Golovchenko, J.A. Ion-beam sculpting at nanometre length scales. Nature 2001, 412, 166–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanunu, M. Nanopores: A journey towards DNA sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Gershow, M.; Stein, D.; Brandin, E.; Golovchenko, J.A. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mat. 2003, 2, 611–615. [Google Scholar] [CrossRef]
- Oukhaled, A.; Bacri, L.; Pastoriza-Gallego, M.; Betton, J.-M.; Pelta, J. Sensing Proteins through Nanopores: Fundamental to Applications. ACS Chem. Biol. 2012, 7, 1935–1949. [Google Scholar] [CrossRef]
- Storm, A.J.; Chen, J.H.; Zandbergen, H.W.; Dekker, C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys. Rev. E 2005, 71, 051903. [Google Scholar] [CrossRef] [Green Version]
- Yusko, E.C.; Bruhn, B.R.; Eggenberger, O.M.; Houghtaling, J.; Rollings, R.C.; Walsh, N.C.; Nandivada, S.; Pindrus, M.; Hall, A.R.; Sept, D.; et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 2017, 12, 360. [Google Scholar] [CrossRef] [Green Version]
- Charron, M.; Briggs, K.; King, S.; Waugh, M.; Tabard-Cossa, V. Precise DNA Concentration Measurements with Nanopores by Controlled Counting. Anal. Chem. 2019, 91, 12228–12237. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Yu, L.; Li, Y.; Qian, G.; Chang, S. Nanopipettes: A potential tool for DNA detection. Analyst 2019, 144, 5037–5047. [Google Scholar] [CrossRef]
- Dekker, C. Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Howorka, S.; Siwy, Z. Nanopore analytics: Sensing of single molecules. Chem. Soc. Rev. 2009, 38, 2360–2384. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Guo, W.; Jiang, L. Biomimetic smart nanopores and nanochannels. Chem. Soc. Rev. 2011, 40, 2385–2401. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.F.; Dekker, C. DNA sequencing with nanopores. Nat. Biotechnol. 2012, 30, 326–328. [Google Scholar] [CrossRef]
- Steinbock, L.J.; Keyser, U.F. Analyzing Single DNA Molecules by Nanopore Translocation. In Nanopore-Based Technology; Gracheva, M.E., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; pp. 135–145. ISBN 978-1-61779-773-6. [Google Scholar]
- Miles, B.N.; Ivanov, A.P.; Wilson, K.A.; Doğan, F.; Japrung, D.; Edel, J.B. Single molecule sensing with solid-state nanopores: Novel materials, methods, and applications. Chem. Soc. Rev. 2012, 42, 15–28. [Google Scholar] [CrossRef]
- Hernández-Ainsa, S.; Keyser, U.F. DNA origami nanopores: An emerging tool in biomedicine. Nanomedicine 2013, 8, 1551–1554. [Google Scholar] [CrossRef]
- Haque, F.; Li, J.; Wu, H.-C.; Liang, X.-J.; Guo, P. Solid-State and Biological Nanopore for Real-Time Sensing of Single Chemical and Sequencing of DNA. Nano Today 2013, 8, 56–74. [Google Scholar] [CrossRef] [Green Version]
- Bell, N.A.W.; Keyser, U.F. Nanopores formed by DNA origami: A review. FEBS Lett. 2014, 588, 3564–3570. [Google Scholar] [CrossRef]
- Makra, I.; Gyurcsányi, R.E. Electrochemical sensing with nanopores: A mini review. Electrochem. Commun. 2014, 43, 55–59. [Google Scholar] [CrossRef] [Green Version]
- Wasfi, A.; Awwad, F.; Ayesh, A.I. Graphene-based nanopore approaches for DNA sequencing: A literature review. Biosens. Bioelectron. 2018, 119, 191–203. [Google Scholar] [CrossRef]
- Varongchayakul, N.; Song, J.; Meller, A.W.; Grinstaff, M. Single-molecule protein sensing in a nanopore: A tutorial. Chem. Soc. Rev. 2018, 47, 8512–8524. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Gao, P.; Ma, Q.; Wang, D.; Xia, F. Biomolecule-Functionalized Solid-State Ion Nanochannels/Nanopores: Features and Techniques. Small 2019, 15, 1804878. [Google Scholar] [CrossRef] [PubMed]
- Fragasso, A.; Schmid, S.; Dekker, C. Comparing Current Noise in Biological and Solid-State Nanopores. ACS Nano 2020, 14, 1338–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanopores; Iqbal, S.M.; Bashir, R. (Eds.) Springer: Boston, MA, USA, 2011; ISBN 978-1-4419-8251-3. [Google Scholar]
- Branton, D.; Deamer, D. Nanopore Sequencing: An Introduction; World Scientific: Singapore, 2019; ISBN 978-981-327-060-2. [Google Scholar]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. TrAC Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Cho, E.J.; Lee, J.-W.; Ellington, A.D. Applications of Aptamers as Sensors. Annu. Rev. Anal. Chem. 2009, 2, 241–264. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Balijepalli, A.K.; Ettedgui, J.; Forstater, J.H.; Wang, H.; Zhang, H.; Robertson, J.W.F. Analytical applications for pore-forming proteins. Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1858, 593–606. [Google Scholar] [CrossRef]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef]
- Song, L.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of Staphylococcal α-Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1865. [Google Scholar] [CrossRef]
- Shi, W.; Friedman, A.K.; Baker, L.A. Nanopore Sensing. Anal. Chem. 2017, 89, 157–188. [Google Scholar] [CrossRef] [Green Version]
- Fahie, M.; Chisholm, C.; Chen, M. Resolved Single-Molecule Detection of Individual Species within a Mixture of anti-Biotin Antibodies Using an Engineered Monomeric Nanopore. ACS Nano 2015, 9, 1089–1098. [Google Scholar] [CrossRef] [Green Version]
- Laszlo, A.H.; Derrington, I.M.; Gundlach, J.H. MspA nanopore as a single-molecule tool: From sequencing to SPRNT. Methods 2016, 105, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Piguet, F.; Ouldali, H.; Pastoriza-Gallego, M.; Manivet, P.; Pelta, J.; Oukhaled, A. Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore. Nat. Commun. 2018, 9, 966. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Cirauqui, N.; Marcaida, M.J.; Buglakova, E.; Duperrex, A.; Radenovic, A.; Dal Peraro, M. Single-molecule sensing of peptides and nucleic acids by engineered aerolysin nanopores. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meervelt, V.; Soskine, M.; Singh, S.; Schuurman-Wolters, G.K.; Wijma, H.J.; Poolman, B.; Maglia, G. Real-Time Conformational Changes and Controlled Orientation of Native Proteins Inside a Protein Nanoreactor. J. Am. Chem. Soc. 2017, 139, 18640–18646. [Google Scholar] [CrossRef] [Green Version]
- Wloka, C.; Van Meervelt, V.; van Gelder, D.; Danda, N.; Jager, N.; Williams, C.P.; Maglia, G. Label-Free and Real-Time Detection of Protein Ubiquitination with a Biological Nanopore. ACS Nano 2017, 11, 4387–4394. [Google Scholar] [CrossRef] [Green Version]
- Haque, F.; Lunn, J.; Fang, H.; Smithrud, D.; Guo, P. Real-time sensing and discrimination of single chemicals using the channel of phi29 DNA packaging nanomotor. ACS Nano 2012, 6, 3251–3261. [Google Scholar] [CrossRef] [Green Version]
- Ji, Z.; Wang, S.; Zhao, Z.; Zhou, Z.; Haque, F.; Guo, P. Fingerprinting of Peptides with a Large Channel of Bacteriophage Phi29 DNA Packaging Motor. Small 2016, 12, 4572–4578. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhao, Z.; Haque, F.; Guo, P. Engineering of protein nanopores for sequencing, chemical or protein sensing and disease diagnosis. Curr. Opin. Biotechnol. 2018, 51, 80–89. [Google Scholar] [CrossRef]
- Franssila, S. Introduction to Microfabrication, 2nd ed.; Wiley: Hoboken, NJ, USA, 2010; ISBN 978-0-470-74983-8. [Google Scholar]
- Storm, A.J.; Chen, J.H.; Ling, X.S.; Zandbergen, H.W.; Dekker, C. Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mat. 2003, 2, 537–540. [Google Scholar] [CrossRef]
- Kennedy, E.; Dong, Z.; Tennant, C.; Timp, G. Reading the primary structure of a protein with 0.07 nm 3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol. 2016, 11, 968–976. [Google Scholar] [CrossRef]
- Graf, M.; Lihter, M.; Thakur, M.; Georgiou, V.; Topolancik, J.; Ilic, B.R.; Liu, K.; Feng, J.; Astier, Y.; Radenovic, A. Fabrication and practical applications of molybdenum disulfide nanopores. Nat. Protoc. 2019, 14, 1130–1168. [Google Scholar] [CrossRef] [PubMed]
- Kwok, H.; Briggs, K.; Tabard-Cossa, V. Nanopore fabrication by controlled dielectric breakdown. PLoS ONE 2014, 9, e92880. [Google Scholar] [CrossRef] [PubMed]
- Yanagi, I.; Akahori, R.; Hatano, T.; Takeda, K. Fabricating nanopores with diameters of sub-1 nm to 3 nm using multilevel pulse-voltage injection. Sci. Rep. 2014, 4, 5000. [Google Scholar] [CrossRef]
- Garaj, S.; Hubbard, W.; Reina, A.; Kong, J.; Branton, D.; Golovchenko, J.A. Graphene as a subnanometre trans-electrode membrane. Nature 2010, 467, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Merchant, C.A.; Healy, K.; Wanunu, M.; Ray, V.; Peterman, N.; Bartel, J.; Fischbein, M.D.; Venta, K.; Luo, Z.; Johnson, A.T.C.; et al. DNA Translocation through Graphene Nanopores. Nano Lett. 2010, 10, 2915–2921. [Google Scholar] [CrossRef] [PubMed]
- Goyal, G.; Lee, Y.B.; Darvish, A.; Ahn, C.W.; Kim, M.J. Hydrophilic and size-controlled graphene nanopores for protein detection. Nanotechnology 2016, 27, 495301. [Google Scholar] [CrossRef] [Green Version]
- Wilson, J.; Sloman, L.; He, Z.; Aksimentiev, A. Graphene Nanopores for Protein Sequencing. Adv. Funct. Mat. 2016, 26, 4830–4838. [Google Scholar] [CrossRef] [Green Version]
- Larkin, J.; Henley, R.; Bell, D.C.; Cohen-Karni, T.; Rosenstein, J.K.; Wanunu, M. Slow DNA Transport through Nanopores in Hafnium Oxide Membranes. ACS Nano 2013, 7, 10121–10128. [Google Scholar] [CrossRef] [Green Version]
- Kudr, J.; Skalickova, S.; Nejdl, L.; Moulick, A.; Ruttkay–Nedecky, B.; Adam, V.; Kizek, R. Fabrication of solid-state nanopores and its perspectives. Electrophoresis 2015, 36, 2367–2379. [Google Scholar] [CrossRef]
- Ando, G.; Hyun, C.; Li, J.; Mitsui, T. Directly Observing the Motion of DNA Molecules near Solid-State Nanopores. ACS Nano 2012, 6, 10090–10097. [Google Scholar] [CrossRef] [Green Version]
- Kubota, T.; Lloyd, K.; Sakashita, N.; Minato, S.; Ishida, K.; Mitsui, T. Clog and Release, and Reverse Motions of DNA in a Nanopore. Polymers 2019, 11, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, A.R.; Scott, A.; Rotem, D.; Mehta, K.K.; Bayley, H.; Dekker, C. Hybrid pore formation by directed insertion of α-haemolysin into solid-state nanopores. Nat. Nanotechnol. 2010, 5, 874–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabello-Aguilar, S.; Balme, S.; Chaaya, A.A.; Bechelany, M.; Balanzat, E.; Janot, J.-M.; Pochat-Bohatier, C.; Miele, P.; Dejardin, P. Slow translocation of polynucleotides and their discrimination by α-hemolysin inside a single track-etched nanopore designed by atomic layer deposition. Nanoscale 2013, 5, 9582–9586. [Google Scholar] [CrossRef] [PubMed]
- Bentin, J.; Balme, S.; Picaud, F. Polynucleotide differentiation using hybrid solid-state nanopore functionalizing with α-hemolysin. Soft Matter 2020, 16, 1002–1010. [Google Scholar] [CrossRef]
- Jovanovic-Talisman, T.; Tetenbaum-Novatt, J.; McKenney, A.S.; Zilman, A.; Peters, R.; Rout, M.P.; Chait, B.T. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 2009, 457, 1023–1027. [Google Scholar] [CrossRef]
- Kowalczyk, S.W.; Kapinos, L.; Blosser, T.R.; Magalhães, T.; van Nies, P.; Lim, R.Y.H.; Dekker, C. Single-molecule transport across an individual biomimetic nuclear pore complex. Nat. Nanotechnol. 2011, 6, 433–438. [Google Scholar] [CrossRef]
- Cressiot, B.; Greive, S.J.; Mojtabavi, M.; Antson, A.A.; Wanunu, M. Thermostable virus portal proteins as reprogrammable adapters for solid-state nanopore sensors. Nat. Commun. 2018, 9, 4652. [Google Scholar] [CrossRef]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Ke, Y. The Beauty and Utility of DNA Origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Martin, T.G.; Rant, U.; Dietz, H. DNA Origami Gatekeepers for Solid-State Nanopores. Angew. Chem. Int. Ed. 2012, 51, 4864–4867. [Google Scholar] [CrossRef]
- Bell, N.A.W.; Engst, C.R.; Ablay, M.; Divitini, G.; Ducati, C.; Liedl, T.; Keyser, U.F. DNA Origami Nanopores. Nano Lett. 2012, 12, 512–517. [Google Scholar] [CrossRef]
- Langecker, M.; Arnaut, V.; Martin, T.G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F.C. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 2012, 338, 932–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Ainsa, S.; Keyser, U.F. DNA origami nanopores: Developments, challenges and perspectives. Nanoscale 2014, 6, 14121–14132. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Seifert, A.; Fertig, N.; Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 2016, 11, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Barati Farimani, A.; Dibaeinia, P.; Aluru, N.R. DNA Origami–Graphene Hybrid Nanopore for DNA Detection. ACS Appl. Mat. Interfaces 2017, 9, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, R.; Pal, S.; Joshi, H.; Rao, A.; Naik, A.; Varma, M.; Chakraborty, B.; Maiti, P.K. DNA Translocation through Hybrid Bilayer Nanopores. J. Phys. Chem. C 2019, 123, 11908–11916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanunu, M.; Meller, A. Chemically Modified Solid-State Nanopores. Nano Lett. 2007, 7, 1580–1585. [Google Scholar] [CrossRef]
- Jonkheijm, P.; Weinrich, D.; Schröder, H.; Niemeyer, C.M.; Waldmann, H. Chemical Strategies for Generating Protein Biochips. Angew. Chem. Int. Ed. 2008, 47, 30. [Google Scholar] [CrossRef]
- Yusko, E.C.; Johnson, J.M.; Majd, S.; Prangkio, P.; Rollings, R.C.; Li, J.; Yang, J.; Mayer, M. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nanotechnol. 2011, 6, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Eggenberger, O.M.; Leriche, G.; Koyanagi, T.; Ying, C.; Houghtaling, J.; Schroeder, T.B.H.; Yang, J.; Li, J.; Hall, A.; Mayer, M. Fluid surface coatings for solid-state nanopores: Comparison of phospholipid bilayers and archaea-inspired lipid monolayers. Nanotechnology 2019, 30, 325504. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Hu, R.; Li, J.; Tong, X.; Diao, J.J.; Yu, D.; Zhao, Q. Non-sticky translocation of bio-molecules through Tween 20-coated solid-state nanopores in a wide pH range. Appl. Phys. Lett. 2016, 109, 143105. [Google Scholar] [CrossRef]
- Shan, Y.P.; Tiwari, P.B.; Krishnakumar, P.; Vlassiouk, I.; Li, W.Z.; Wang, X.W.; Darici, Y.; Lindsay, S.M.; Wang, H.D.; Smirnov, S.; et al. Surface modification of graphene nanopores for protein translocation. Nanotechnology 2013, 24, 495102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roman, J.; Jarroux, N.; Patriarche, G.; Français, O.; Pelta, J.; Le Pioufle, B.; Bacri, L. Functionalized Solid-State Nanopore Integrated in a Reusable Microfluidic Device for a Better Stability and Nanoparticle Detection. ACS Appl. Mat. Interfaces 2017, 9, 41634–41640. [Google Scholar] [CrossRef] [PubMed]
- Lepoitevin, M.; Ma, T.; Bechelany, M.; Janot, J.-M.; Balme, S. Functionalization of single solid state nanopores to mimic biological ion channels: A review. Adv. Colloid Interface Sci. 2017, 250, 195–213. [Google Scholar] [CrossRef]
- Eggenberger, O.M.; Ying, C.; Mayer, M. Surface coatings for solid-state nanopores. Nanoscale 2019, 11, 19636–19657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atomic Layer Deposition: An Overview/Chemical Reviews. Available online: https://pubs.acs.org/doi/10.1021/cr900056b (accessed on 8 May 2020).
- Hampden-Smith, M.J.; Kodas, T.T. Chemical vapor deposition of metals: Part 1. An overview of CVD processes. Chem. Vapor Depos. 1995, 1, 8–23. [Google Scholar] [CrossRef]
- Asatekin, A.; Gleason, K.K. Polymeric Nanopore Membranes for Hydrophobicity-Based Separations by Conformal Initiated Chemical Vapor Deposition. Nano Lett. 2011, 11, 677–686. [Google Scholar] [CrossRef]
- Chen, P.; Mitsui, T.; Farmer, D.B.; Golovchenko, J.; Gordon, R.G.; Branton, D. Atomic Layer Deposition to Fine-Tune the Surface Properties and Diameters of Fabricated Nanopores. Nano Lett. 2004, 4, 1333–1337. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Fu, Q.; Wang, X.; Kong, D.; Sheng, Q.; Wang, Y.; Chen, Q.; Xue, J. Atomic Layer Deposition Modified Track-Etched Conical Nanochannels for Protein Sensing. Anal. Chem. 2015, 87, 8227–8233. [Google Scholar] [CrossRef]
- Chou, Y.-C.; Masih Das, P.; Monos, D.S.; Drndić, M. Lifetime and Stability of Silicon Nitride Nanopores and Nanopore Arrays for Ionic Measurements. ACS Nano 2020, 14, 6715–6728. [Google Scholar] [CrossRef]
- Yamazaki, H.; Hu, R.; Zhao, Q.; Wanunu, M. Photothermally Assisted Thinning of Silicon Nitride Membranes for Ultrathin Asymmetric Nanopores. ACS Nano 2018, 12, 12472–12481. [Google Scholar] [CrossRef]
- Hu, R.; Diao, J.; Li, J.; Tang, Z.; Li, X.; Leitz, J.; Long, J.; Liu, J.; Yu, D.; Zhao, Q. Intrinsic and membrane-facilitated α-synuclein oligomerization revealed by label-free detection through solid-state nanopores. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Schneider, G.F.; Xu, Q.; Hage, S.; Luik, S.; Spoor, J.N.H.; Malladi, S.; Zandbergen, H.; Dekker, C. Tailoring the hydrophobicity of graphene for its use as nanopores for DNA translocation. Nat. Commun. 2013, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Umehara, S.; Pourmand, N.; Webb, C.D.; Davis, R.W.; Yasuda, K.; Karhanek, M. Current Rectification with Poly-l-Lysine-Coated Quartz Nanopipettes. Nano Lett. 2006, 6, 2486–2492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Actis, P.; Rogers, A.; Nivala, J.; Vilozny, B.; Seger, R.A.; Jejelowo, O.; Pourmand, N. Reversible thrombin detection by aptamer functionalized STING sensors. Biosens. Bioelectron. 2011, 26, 4503–4507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Actis, P.; Jejelowo, O.; Pourmand, N. UltraSensitive Mycotoxin Detection by STING Sensors. Biosens. Bioelectron. 2010, 26, 333–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alem, H.; Blondeau, F.; Glinel, K.; Demoustier-Champagne, S.; Jonas, A.M. Layer-by-Layer Assembly of Polyelectrolytes in Nanopores. Macromolecules 2007, 40, 3366–3372. [Google Scholar] [CrossRef]
- Ali, M.; Yameen, B.; Cervera, J.; Ramírez, P.; Neumann, R.; Ensinger, W.; Knoll, W.; Azzaroni, O. Layer-by-Layer Assembly of Polyelectrolytes into Ionic Current Rectifying Solid-State Nanopores: Insights from Theory and Experiment. J. Am. Chem. Soc. 2010, 132, 8338–8348. [Google Scholar] [CrossRef]
- Ma, T.; Gaigalas, P.; Lepoitevin, M.; Plikusiene, I.; Bechelany, M.; Janot, J.-M.; Balanzat, E.; Balme, S. Impact of Polyelectrolyte Multilayers on the Ionic Current Rectification of Conical Nanopores. Langmuir 2018, 34, 3405–3412. [Google Scholar] [CrossRef]
- Lepoitevin, M.; Jamilloux, B.; Bechelany, M.; Balanzat, E.; Janot, J.-M.; Balme, S. Fast and reversible functionalization of a single nanopore based on layer-by-layer polyelectrolyte self-assembly for tuning current rectification and designing sensors. RSC Adv. 2016, 6, 32228–32233. [Google Scholar] [CrossRef]
- Blundell, E.L.C.J.; Mayne, L.J.; Lickorish, M.; Christie, S.D.R.; Platt, M. Protein detection using tunable pores: Resistive pulses and current rectification. Faraday Discuss. 2016, 193, 487–505. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Xu, X.; Wang, P.; Chen, L.; Jin, Y. The facile surface chemical modification of a single glass nanopore and its use in the nonenzymatic detection of uric acid. Chem. Commun. 2015, 51, 1914–1917. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Gatterdam, V.; Wieneke, R.; Tampé, R.; Rant, U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol. 2012, 7, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Siwy, Z.; Trofin, L.; Kohli, P.; Baker, L.A.; Trautmann, C.; Martin, C.R. Protein Biosensors Based on Biofunctionalized Conical Gold Nanotubes. J. Am. Chem. Soc. 2005, 127, 5000–5001. [Google Scholar] [CrossRef] [PubMed]
- Emilsson, G.; Xiong, K.; Sakiyama, Y.; Malekian, B.; Gagnér, V.A.; Schoch, R.L.; Lim, R.Y.H.; Dahlin, A.B. Polymer brushes in solid-state nanopores form an impenetrable entropic barrier for proteins. Nanoscale 2018, 10, 4663–4669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emilsson, G.; Sakiyama, Y.; Malekian, B.; Xiong, K.; Adali-Kaya, Z.; Lim, R.Y.H.; Dahlin, A.B. Gating Protein Transport in Solid State Nanopores by Single Molecule Recognition. ACS Cent. Sci. 2018, 4, 1007–1014. [Google Scholar] [CrossRef]
- Guo, W.; Xia, H.; Xia, F.; Hou, X.; Cao, L.; Wang, L.; Xue, J.; Zhang, G.; Song, Y.; Zhu, D.; et al. Current Rectification in Temperature-Responsive Single Nanopores. ChemPhysChem 2010, 11, 859–864. [Google Scholar] [CrossRef]
- Schroeder, T.B.H.; Houghtaling, J.; Wilts, B.D.; Mayer, M. It’s Not a Bug, It’s a Feature: Functional Materials in Insects. Adv. Mat. 2018, 30, 1705322. [Google Scholar] [CrossRef] [Green Version]
- Brzoska, J.B.; Azouz, I.B.; Rondelez, F. Silanization of Solid Substrates: A Step Toward Reproducibility. Langmuir 1994, 10, 4367–4373. [Google Scholar] [CrossRef]
- To, T.D.; Nguyen, A.T.; Phan, K.N.T.; Truong, A.T.T.; Doan, T.C.D.; Dang, C.M. Modification of silicon nitride surfaces with GOPES and APTES for antibody immobilization: Computational and experimental studies. Adv. Nat. Sci. Nanosci. Nanotechnol. 2015, 6, 045006. [Google Scholar] [CrossRef]
- Nilsson, J.; Lee, J.R.I.; Ratto, T.V.; Létant, S.E. Localized Functionalization of Single Nanopores. Adv. Mat. 2006, 18, 427–431. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, S.M.; Akin, D.; Bashir, R. Solid-state nanopore channels with DNA selectivity. Nat. Nanotechnol. 2007, 2, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Gao, C.; Gu, L.-Q. Capturing Single Molecules of Immunoglobulin and Ricin with an Aptamer-Encoded Glass Nanopore. Anal. Chem. 2009, 81, 6649–6655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mussi, V.; Fanzio, P.; Repetto, L.; Firpo, G.; Stigliani, S.; Tonini, G.P.; Valbusa, U. “DNA-Dressed NAnopore” for complementary sequence detection. Biosens. Bioelectron. 2011, 29, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-C.; Gao, M.-J.; Chen, W.; Hu, X.-Y.; Song, L.-B.; Liu, B.; Zhao, Y.-D. pH-modulated ion-current rectification in a cysteine-functionalized glass nanopipette. Electrochem. Commun. 2018, 97, 6–10. [Google Scholar] [CrossRef]
- Liebes-Peer, Y.; Rapaport, H.; Ashkenasy, N. Amplification of Single Molecule Translocation Signal Using β-Strand Peptide Functionalized Nanopores. ACS Nano 2014, 8, 6822–6832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, G.; Chai, H.; Zhao, Y.-D.; Yu, L.; Chen, W. Detection of alkaline phosphatase activity with a functionalized nanopipette. Electrochem. Commun. 2019, 99, 71–74. [Google Scholar] [CrossRef]
- Zhang, L.-X.; Cai, S.-L.; Zheng, Y.-B.; Cao, X.-H.; Li, Y.-Q. Smart Homopolymer Modification to Single Glass Conical Nanopore Channels: Dual-Stimuli-Actuated Highly Efficient Ion Gating. Adv. Funct. Mat. 2011, 21, 2103–2107. [Google Scholar] [CrossRef]
- Fu, Y.; Tokuhisa, H.; Baker, L.A. Nanopore DNA sensors based on dendrimer-modified nanopipettes. Chem. Commun. 2009, 4877–4879. [Google Scholar] [CrossRef]
- Lee, S.B.; Mitchell, D.T.; Trofin, L.; Nevanen, T.K.; Söderlund, H.; Martin, C.R. Antibody-Based Bio-Nanotube Membranes for Enantiomeric Drug Separations. Science 2002, 296, 2198–2200. [Google Scholar] [CrossRef]
- Wang, G.; Bohaty, A.K.; Zharov, I.; White, H.S. Photon Gated Transport at the Glass Nanopore Electrode. J. Am. Chem. Soc. 2006, 128, 13553–13558. [Google Scholar] [CrossRef]
- Ananth, A.; Genua, M.; Aissaoui, N.; Díaz, L.; Eisele, N.B.; Frey, S.; Dekker, C.; Richter, R.P.; Görlich, D. Reversible Immobilization of Proteins in Sensors and Solid-State Nanopores. Small 2018, 14, 1703357. [Google Scholar] [CrossRef] [PubMed]
- Bouchet, A.; Descamps, E.; Mailley, P.; Livache, T.; Chatelain, F.; Haguet, V. Contactless Electrofunctionalization of a Single Pore. Small 2009, 5, 2297–2303. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Pham, P.; Haguet, V.; Sauter-Starace, F.; Leroy, L.; Roget, A.; Descamps, E.; Bouchet, A.; Buhot, A.; Mailley, P.; et al. Polarization-Induced Local Pore-Wall Functionalization for Biosensing: From Micropore to Nanopore. Anal. Chem. 2012, 84, 3254–3261. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.; Voci, S.; Pham, P.; Leroy, L.; Maziz, A.; Descamps, L.; Kuhn, A.; Mailley, P.; Livache, T.; Buhot, A.; et al. Enhanced Bipolar Electrochemistry at Solid-State Micropores: Demonstration by Wireless Electrochemiluminescence Imaging. Anal. Chem. 2019, 91, 8900–8907. [Google Scholar] [CrossRef]
- Bouchet-Spinelli, A.; Descamps, E.; Liu, J.; Ismail, A.; Pham, P.; Chatelain, F.; Leïchlé, T.; Leroy, L.; Marche, P.N.; Raillon, C.; et al. Polarization Induced Electro-Functionalization of Pore Walls: A Contactless Technology. Biosensors 2019, 9, 121. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Si, Z.; Li, Y.; Wang, D.; Wu, X.; Gao, P.; Xia, F. Functional solid-state nanochannels for biochemical sensing. TrAC Trends Anal. Chem. 2019, 115, 174–186. [Google Scholar] [CrossRef]
- Clarke, R.W.; White, S.S.; Zhou, D.; Ying, L.; Klenerman, D. Trapping of Proteins under Physiological Conditions in a Nanopipette. Angew. Chem. Int. Ed. 2005, 44, 3747–3750. [Google Scholar] [CrossRef]
- Steinbock, L.J.; Otto, O.; Chimerel, C.; Gornall, J.; Keyser, U.F. Detecting DNA Folding with Nanocapillaries. Nano Lett. 2010, 10, 2493–2497. [Google Scholar] [CrossRef]
- Steinbock, L.J.; Steinbock, J.F.; Radenovic, A. Controllable Shrinking and Shaping of Glass Nanocapillaries under Electron Irradiation. Nano Lett. 2013, 13, 1717–1723. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Bell, N.A.W.; Hernández-Ainsa, S.; Thacker, V.V.; Thackray, A.M.; Bujdoso, R.; Keyser, U.F. Single Protein Molecule Detection by Glass Nanopores. ACS Nano 2013, 7, 4129–4134. [Google Scholar] [CrossRef]
- Terejánszky, P.; Makra, I.; Fürjes, P.; Gyurcsányi, R.E. Calibration-Less Sizing and Quantitation of Polymeric Nanoparticles and Viruses with Quartz Nanopipets. Anal. Chem. 2014, 86, 4688–4697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sze, J.Y.Y.; Kumar, S.; Ivanov, A.P.; Oh, S.-H.; Edel, J.B. Fine tuning of nanopipettes using atomic layer deposition for single molecule sensing. Analyst 2015, 140, 4828–4834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youn, Y.; Lee, C.; Kim, J.H.; Chang, Y.W.; Kim, D.Y.; Yoo, K.-H. Selective Detection of Single-Stranded DNA Molecules Using a Glass Nanocapillary Functionalized with DNA. Anal. Chem. 2016, 88, 688–694. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, B.; Wayment, J.R.; Harris, J.M.; White, H.S. Electrostatic-Gated Transport in Chemically Modified Glass Nanopore Electrodes. J. Am. Chem. Soc. 2006, 128, 7679–7686. [Google Scholar] [CrossRef] [PubMed]
- Bulushev, R.D.; Marion, S.; Petrova, E.; Davis, S.J.; Maerkl, S.J.; Radenovic, A. Single Molecule Localization and Discrimination of DNA–Protein Complexes by Controlled Translocation Through Nanocapillaries. Nano Lett. 2016, 16, 7882–7890. [Google Scholar] [CrossRef] [Green Version]
- Vitol, E.A.; Orynbayeva, Z.; Bouchard, M.J.; Azizkhan-Clifford, J.; Friedman, G.; Gogotsi, Y. In Situ Intracellular Spectroscopy with Surface Enhanced Raman Spectroscopy (SERS)-Enabled Nanopipettes. ACS Nano 2009, 3, 3529–3536. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Melaine, F.; Roupioz, Y.; Buhot, A. Gold Nanoparticles Surface Plasmon Resonance Enhanced Signal for the Detection of Small Molecules on Split-Aptamer Microarrays. Microarrays 2015, 4, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Melaine, F.; Coilhac, C.; Roupioz, Y.; Buhot, A. A nanoparticle-based thermo-dynamic aptasensor for small molecule detection. Nanoscale 2016, 8, 16947–16954. [Google Scholar] [CrossRef]
- Daniel, C.; Mélaïne, F.; Roupioz, Y.; Livache, T.; Buhot, A. Real time monitoring of thrombin interactions with its aptamers: Insights into the sandwich complex formation. Biosens. Bioelectron. 2013, 40, 186–192. [Google Scholar] [CrossRef]
- Daniel, C.; Roupioz, Y.; Gasparutto, D.; Livache, T.; Buhot, A. Solution-Phase vs Surface-Phase Aptamer-Protein Affinity from a Label-Free Kinetic Biosensor. PLoS ONE 2013, 8, e75419. [Google Scholar] [CrossRef] [Green Version]
- Shim, J.W.; Gu, L.-Q. Encapsulating a Single G-Quadruplex Aptamer in a Protein Nanocavity. J. Phys. Chem. B 2008, 112, 8354–8360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meervelt, V.; Soskine, M.; Maglia, G. Detection of Two Isomeric Binding Configurations in a Protein–Aptamer Complex with a Biological Nanopore. ACS Nano 2014, 8, 12826–12835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnaut, V.; Langecker, M.; Simmel, F.C. Nanopore Force Spectroscopy of Aptamer–Ligand Complexes. Biophys. J. 2013, 105, 1199–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winters-Hilt, S.; Davis, A.; Amin, I.; Morales, E. Nanopore current transduction analysis of protein binding to non-terminal and terminal DNA regions: Analysis of transcription factor binding, retroviral DNA terminus dynamics, and retroviral integrase-DNA binding. BMC Bioinform. 2007, 8, S10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, K.; Amin, I.; Morales, E.; Winters-Hilt, S. Preliminary nanopore cheminformatics analysis of aptamer-target binding strength. BMC Bioinform. 2007, 8, S11. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.-Q.; Wook Shim, J. Single molecule sensing by nanopores and nanopore devices. Analyst 2010, 135, 441–451. [Google Scholar] [CrossRef] [Green Version]
- Renner, S.; Geltinger, S.; Simmel, F.C. Nanopore Translocation and Force Spectroscopy Experiments in Microemulsion Droplets. Small 2010, 6, 190–194. [Google Scholar] [CrossRef]
- Shim, J.; Gu, L.-Q. Single-molecule investigation of G-quadruplex using a nanopore sensor. Methods 2012, 57, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Shim, J.W.; Tan, Q.; Gu, L.-Q. Single-molecule detection of folding and unfolding of the G-quadruplex aptamer in a nanopore nanocavity. Nucleic Acids Res 2009, 37, 972–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmood, M.A.I.; Ali, W.; Adnan, A.; Iqbal, S.M. 3D Structural Integrity and Interactions of Single-Stranded Protein-Binding DNA in a Functionalized Nanopore. J. Phys. Chem. B 2014, 118, 5799–5806. [Google Scholar] [CrossRef] [PubMed]
- Billinge, E.R.; Broom, M.; Platt, M. Monitoring Aptamer–Protein Interactions Using Tunable Resistive Pulse Sensing. Anal. Chem. 2014, 86, 1030–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.-P.; Cao, J.; Nie, X.-G.; Wang, S.-S.; Wang, C.; Xia, X.-H. Label-free monitoring of the thrombin–aptamer recognition reaction using an array of nanochannels coupled with electrochemical detection. Electrochem. Commun. 2017, 81, 5–9. [Google Scholar] [CrossRef]
- Mohammad, M.M.; Iyer, R.; Howard, K.R.; McPike, M.P.; Borer, P.N.; Movileanu, L. Engineering a Rigid Protein Tunnel for Biomolecular Detection. J. Am. Chem. Soc. 2012, 134, 9521–9531. [Google Scholar] [CrossRef] [Green Version]
- Ying, Y.-L.; Wang, H.-Y.; Sutherland, T.C.; Long, Y.-T. Monitoring of an ATP-Binding Aptamer and its Conformational Changes Using an α-Hemolysin Nanopore. Small 2011, 7, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, J.; Ying, Y.-L.; Tian, H.; Long, Y.-T. Single molecule analysis of light-regulated RNA:spiropyran interactions. Chem. Sci. 2014, 5, 2642–2646. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Song, Z.-Y.; Zhang, H.-S.; Chen, S.-P. Single-molecule analysis of lead (II)-binding aptamer conformational changes in an α-hemolysin nanopore, and sensitive detection of lead (II). Microchim. Acta 2016, 183, 1003–1010. [Google Scholar] [CrossRef]
- Mayne, L.; Lin, C.-Y.; Christie, S.D.R.; Siwy, Z.S.; Platt, M. The Design and Characterization of Multifunctional Aptamer Nanopore Sensors. ACS Nano 2018, 12, 4844–4852. [Google Scholar] [CrossRef] [Green Version]
- Kawano, R.; Osaki, T.; Sasaki, H.; Takinoue, M.; Yoshizawa, S.; Takeuchi, S. Rapid Detection of a Cocaine-Binding Aptamer Using Biological Nanopores on a Chip. J. Am. Chem. Soc. 2011, 133, 8474–8477. [Google Scholar] [CrossRef]
- Sze, J.Y.Y.; Ivanov, A.P.; Cass, A.E.G.; Edel, J.B. Single molecule multiplexed nanopore protein screening in human serum using aptamer modified DNA carriers. Nat. Commun. 2017, 8, 1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zhang, K.; Liu, G.; Liu, M.; Liu, Y.; Li, J. Label-Free Nanopore Proximity Bioassay for Platelet-Derived Growth Factor Detection. Anal. Chem. 2015, 87, 5677–5682. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, L.; Li, Y.; Xie, J.; Wu, H.-C. A Universal Strategy for Aptamer-Based Nanopore Sensing through Host–Guest Interactions inside α-Hemolysin. Angew. Chem. Int. Ed. 2015, 54, 7568–7571. [Google Scholar] [CrossRef] [PubMed]
- Niedzwiecki, D.J.; Iyer, R.; Borer, P.N.; Movileanu, L. Sampling a Biomarker of the Human Immunodeficiency Virus across a Synthetic Nanopore. ACS Nano 2013, 7, 3341–3350. [Google Scholar] [CrossRef] [Green Version]
- Zeng, T.; Li, T.; Li, Y.; Liu, L.; Wang, X.; Liu, Q.; Zhao, Y.; Wu, H.-C. DNA-based detection of mercury (II) ions through characteristic current signals in nanopores with high sensitivity and selectivity. Nanoscale 2014, 6, 8579–8584. [Google Scholar] [CrossRef]
- Zhang, S.; Bao, A.; Sun, T.; Wang, E.; Wang, J. PEI/Zr4+-coated nanopore for selective and sensitive detection of ATP in combination with single-walled carbon nanotubes. Biosens. Bioelectron. 2015, 63, 287–293. [Google Scholar] [CrossRef]
- Kawano, R.; Osaki, T.; Sasaki, H.; Takinoue, M.; Yoshizawa, S.; Takeuchi, S. 25 second cocaine sensing by membrane protein channel integrated in a microfluidic device. In Proceedings of the 2011 IEEE 24th International Conference on Micro Electro Mechanical Systems, Cancun, Mexico, 23–27 January 2011; pp. 1333–1336. [Google Scholar]
- Rauf, S.; Zhang, L.; Ali, A.; Liu, Y.; Li, J. Label-Free Nanopore Biosensor for Rapid and Highly Sensitive Cocaine Detection in Complex Biological Fluids. ACS Sens. 2017, 2, 227–234. [Google Scholar] [CrossRef]
- Nobukawa, A.; Osaki, T.; Tonooka, T.; Morimoto, Y.; Takeuchi, S. Electrical detection of pesticide vapors by biological nanopores with DNA aptamers. In Proceedings of the 2015 28th IEEE International Conference on Micro Electro Mechanical Systems (MEMS), Estoril, Portugal, 18–22 January 2015; pp. 596–599. [Google Scholar]
- Fujii, S.; Misawa, N.; Kamiya, K.; Osaki, T.; Takeuchi, S. Breathable fabric meets a lipid bilayer system for rapid vapor detection. In Proceedings of the 2018 IEEE Micro Electro Mechanical Systems (MEMS), Belfast, Ireland, 21–25 January 2018; pp. 276–277. [Google Scholar]
- Fujii, S.; Nobukawa, A.; Osaki, T.; Morimoto, Y.; Kamiya, K.; Misawa, N.; Takeuchi, S. Pesticide vapor sensing using an aptamer, nanopore, and agarose gel on a chip. Lab Chip 2017, 17, 2421–2425. [Google Scholar] [CrossRef]
- Bell, N.A.W.; Keyser, U.F. Specific Protein Detection Using Designed DNA Carriers and Nanopores. J. Am. Chem. Soc. 2015, 137, 2035–2041. [Google Scholar] [CrossRef]
- Beamish, E.; Tabard-Cossa, V.; Godin, M. Identifying Structure in Short DNA Scaffolds Using Solid-State Nanopores. ACS Sens. 2017, 2, 1814–1820. [Google Scholar] [CrossRef]
- Kong, J.; Zhu, J.; Chen, K.; Keyser, U.F. Specific Biosensing Using DNA Aptamers and Nanopores. Adv. Funct. Mat. 2019, 29, 1807555. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Lim, M.-C.; Ryu, H.; Shim, J.; Kim, S.M.; Kim, Y.-R.; Jeon, T.-J. Nanopore based detection of Bacillus thuringiensis HD-73 spores using aptamers and versatile DNA hairpins. Nanoscale 2018, 10, 11955–11961. [Google Scholar] [CrossRef] [PubMed]
- Billinge, E.R.; Platt, M. Multiplexed, label-free detection of biomarkers using aptamers and Tunable Resistive Pulse Sensing (AptaTRPS). Biosens. Bioelectron. 2015, 68, 741–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, D.; Li, Z.; Liu, L.; Ai, S.; Zhang, S. Ultrasensitive Detection of Cancer Cells Combining Enzymatic Signal Amplification with an Aerolysin Nanopore. Anal. Chem. 2018, 90, 1029–1034. [Google Scholar] [CrossRef]
- Platt, M.; Willmott, G.R.; Lee, G.U. Resistive Pulse Sensing of Analyte-Induced Multicomponent Rod Aggregation Using Tunable Pores. Small 2012, 8, 2436–2444. [Google Scholar] [CrossRef] [Green Version]
- Billinge, E.R.; Platt, M. Aptamer based dispersion assay using tunable resistive pulse sensing (TRPS). Anal. Methods 2015, 7, 8534–8538. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.P.; Ivanov, A.B.; Edel, J. Selective single molecule nanopore sensing of proteins using DNA aptamer-functionalised gold nanoparticles. Chem. Sci. 2017, 8, 3905–3912. [Google Scholar] [CrossRef] [Green Version]
- Alsager, O.A.; Kumar, S.; Willmott, G.R.; McNatty, K.P.; Hodgkiss, J.M. Small molecule detection in solution via the size contraction response of aptamer functionalized nanoparticles. Biosens. Bioelectron. 2014, 57, 262–268. [Google Scholar] [CrossRef]
- He, F.; Liang, L.; Zhou, S.; Xie, W.; He, S.; Wang, Y.; Tlili, C.; Tong, S.; Wang, D. Label-Free Sensitive Detection of Microcystin-LR via Aptamer-Conjugated Gold Nanoparticles Based on Solid-State Nanopores. Langmuir 2018, 34, 14825–14833. [Google Scholar] [CrossRef]
- Healey, M.J.; Sivakumaran, M.; Platt, M. Rapid quantification of prion proteins using resistive pulse sensing. Analyst 2020, 145, 2595–2601. [Google Scholar] [CrossRef]
- Tang, H.; Wang, H.; Yang, C.; Zhao, D.; Qian, Y.; Li, Y. Nanopore-based Strategy for Selective Detection of Single Carcinoembryonic Antigen (CEA) Molecules. Anal. Chem. 2020, 92, 3042–3049. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Liu, L.; Wang, Y.; Xi, D.; Zhang, S. Unambiguous Discrimination of Multiple Protein Biomarkers by Nanopore Sensing with Double-Stranded DNA-Based Probes. Anal. Chem. 2020, 92, 1730–1737. [Google Scholar] [CrossRef]
- Siwy, Z.; Heins, E.; Harrell, C.C.; Kohli, P.; Martin, C.R. Conical-Nanotube Ion-Current Rectifiers: The Role of Surface Charge. J. Am. Chem. Soc. 2004, 126, 10850–10851. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Feng, Y.; Su, J.; Nie, J.; Cao, L.; Mao, L.; Jiang, L.; Guo, W. On the Origin of Ionic Rectification in DNA-Stuffed Nanopores: The Breaking and Retrieving Symmetry. J. Am. Chem. Soc. 2017, 139, 18739–18746. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zeng, S.; Li, S.; Zhang, Z.; Zhang, S.-L. On Rectification of Ionic Current in Nanopores. Anal. Chem. 2019, 91, 14597–14604. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.-L.; Cao, S.-H.; Zheng, Y.-B.; Zhao, S.; Yang, J.-L.; Li, Y.-Q. Surface charge modulated aptasensor in a single glass conical nanopore. Biosens. Bioelectron. 2015, 71, 37–43. [Google Scholar] [CrossRef]
- Ali, M.; Nasir, S.; Ensinger, W. Bioconjugation-induced ionic current rectification in aptamer-modified single cylindrical nanopores. Chem. Commun. 2015, 51, 3454–3457. [Google Scholar] [CrossRef]
- Zhang, S.; Chai, H.; Cheng, K.; Song, L.; Chen, W.; Yu, L.; Lu, Z.; Liu, B.; Zhao, Y.-D. Ultrasensitive and regenerable nanopore sensing based on target induced aptamer dissociation. Biosens. Bioelectron. 2020, 152, 112011. [Google Scholar] [CrossRef]
- Das, N.; Ray, R.; Ray, S.; Roychaudhuri, C. Intelligent Quantification of Picomolar Protein Concentration in Serum by Functionalized Nanopores. IEEE Sens. J. 2018, 18, 10183–10191. [Google Scholar] [CrossRef]
- Zhao, X.-P.; Zhou, Y.; Zhang, Q.-W.; Yang, D.-R.; Wang, C.; Xia, X.-H. Nanochannel–Ion Channel Hybrid Device for Ultrasensitive Monitoring of Biomolecular Recognition Events. Anal. Chem. 2019, 91, 1185–1193. [Google Scholar] [CrossRef]
- Varga, M.; Bérczes, Z.; Illés, L.; Sáfrány, G.; Bársony, I.; Fürjes, P.; Gyurcsányi, R.E.; Jágerszki, G. Fluidically and electrically integrated solid state nanopore arrays for biochemical sensing. In Proceedings of the 2014 IEEE SENSORS, Valencia, Spain, 2–5 November 2014; pp. 870–872. [Google Scholar]
- Gu, L.-Q.; Ding, S.; Gao, C. Aptamer-encoded nanopore for ultrasensitive detection of bioterrorist agent ricin at single-molecule resolution. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 3–6 September 2009; pp. 6699–6702. [Google Scholar] [CrossRef] [Green Version]
- Abelow, A.E.; Schepelina, O.; White, R.J.; Vallée-Bélisle, A.; Plaxco, K.W.; Zharov, I. Biomimetic glass nanopores employing aptamer gates responsive to a small molecule. Chem. Commun. 2010, 46, 7984–7986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Hou, J.; Zhang, H.; Tian, Y.; Jiang, L. Single Nanochannel-Aptamer-Based Biosensor for Ultrasensitive and Selective Cocaine Detection. ACS Appl. Mat. Interfaces 2018, 10, 2033–2039. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.; Giselbrecht, S.; Rapp, B.E.; Hirtz, M.; Niemeyer, C.M. Advances in DNA-directed immobilization. Curr. Opin. Chem. Biol. 2014, 18, 8–15. [Google Scholar] [CrossRef]
- Liu, N.; Jiang, Y.; Zhou, Y.; Xia, F.; Guo, W.; Jiang, L. Two-Way Nanopore Sensing of Sequence-Specific Oligonucleotides and Small-Molecule Targets in Complex Matrices Using Integrated DNA Supersandwich Structures. Angew. Chem. Int. Ed. 2013, 52, 2007–2011. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, L.; Xu, X.; Liu, S. Quantitative Detection of Potassium Ions and Adenosine Triphosphate via a Nanochannel-Based Electrochemical Platform Coupled with G-Quadruplex Aptamers. Anal. Chem. 2014, 86, 10741–10748. [Google Scholar] [CrossRef]
- Liu, N.; Yang, Z.; Lou, X.; Wei, B.; Zhang, J.; Gao, P.; Hou, R.; Xia, F. Nanopore-Based DNA-Probe Sequence-Evolution Method Unveiling Characteristics of Protein–DNA Binding Phenomena in a Nanoscale Confined Space. Anal. Chem. 2015, 87, 4037–4041. [Google Scholar] [CrossRef]
- Rotem, D.; Jayasinghe, L.; Salichou, M.; Bayley, H. Protein Detection by Nanopores Equipped with Aptamers. J. Am. Chem. Soc. 2012, 134, 2781–2787. [Google Scholar] [CrossRef]
- Soskine, M.; Biesemans, A.; Moeyaert, B.; Cheley, S.; Bayley, H.; Maglia, G. An Engineered ClyA Nanopore Detects Folded Target Proteins by Selective External Association and Pore Entry. Nano Lett. 2012, 12, 4895–4900. [Google Scholar] [CrossRef] [Green Version]
- Hanif, S.; Liu, H.-L.; Ahmed, S.A.; Yang, J.-M.; Zhou, Y.; Pang, J.; Ji, L.-N.; Xia, X.-H.; Wang, K. Nanopipette-Based SERS Aptasensor for Subcellular Localization of Cancer Biomarker in Single Cells. Anal. Chem. 2017, 89, 9911–9917. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, N.; Guo, W.; Xia, F.; Jiang, L. Highly-Efficient Gating of Solid-State Nanochannels by DNA Supersandwich Structure Containing ATP Aptamers: A Nanofluidic IMPLICATION Logic Device. J. Am. Chem. Soc. 2012, 134, 15395–15401. [Google Scholar] [CrossRef]
- Acar, E.T.; Buchsbaum, S.F.; Combs, C.; Fornasiero, F.; Siwy, Z.S. Biomimetic potassium-selective nanopores. Sci. Adv. 2019, 5, eaav2568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Kong, X.-Y.; Xie, G.; Xiao, K.; Zhang, Z.; Wen, L.; Jiang, L. Adenosine-Activated Nanochannels Inspired by G-Protein-Coupled Receptors. Small 2016, 12, 1854–1858. [Google Scholar] [CrossRef] [PubMed]
- Kawano, R. Synthetic Ion Channels and DNA Logic Gates as Components of Molecular Robots. Chem. Phys. Chem. 2018, 19, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Graf, M.; Liu, K.; Ovchinnikov, D.; Dumcenco, D.; Heiranian, M.; Nandigana, V.; Aluru, N.R.; Kis, A.; Radenovic, A. Single-layer MoS 2 nanopores as nanopower generators. Nature 2016, 536, 197–200. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Jin, W.; Xu, N. Two-Dimensional-Material Membranes: A New Family of High-Performance Separation Membranes. Angew. Chem. Int. Ed. 2016, 55, 13384–13397. [Google Scholar] [CrossRef]
- Wang, L.; Boutilier, M.S.H.; Kidambi, P.R.; Jang, D.; Hadjiconstantinou, N.G.; Karnik, R. Fundamental transport mechanisms, fabrication and potential applications of nanoporous atomically thin membranes. Nat. Nanotechnol. 2017, 12, 509–522. [Google Scholar] [CrossRef]
- Surwade, S.P.; Smirnov, S.N.; Vlassiouk, I.V.; Unocic, R.R.; Veith, G.M.; Dai, S.; Mahurin, S.M. Water desalination using nanoporous single-layer graphene. Nat. Nanotechnol. 2015, 10, 459–464. [Google Scholar] [CrossRef]
- Heiranian, M.; Farimani, A.B.; Aluru, N.R. Water desalination with a single-layer MoS 2 nanopore. Nat. Commun. 2015, 6, 8616. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Yang, X.; Liang, L.; Gao, Y.; Cheng, H.; Li, X.; Zou, M.; Ma, R.; Yuan, Q.; Duan, X. Large-area graphene-nanomesh/carbon-nanotube hybrid membranes for ionic and molecular nanofiltration. Science 2019, 364, 1057–1062. [Google Scholar] [CrossRef]
- Dervin, S.; Dionysiou, D.D.; Pillai, S.C. 2D nanostructures for water purification: Graphene and beyond. Nanoscale 2016, 8, 15115–15131. [Google Scholar] [CrossRef]
- Zhang, Z.; Wen, L.; Jiang, L. Bioinspired smart asymmetric nanochannel membranes. Chem. Soc. Rev. 2018, 47, 322–356. [Google Scholar] [CrossRef] [PubMed]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reynaud, L.; Bouchet-Spinelli, A.; Raillon, C.; Buhot, A. Sensing with Nanopores and Aptamers: A Way Forward. Sensors 2020, 20, 4495. https://doi.org/10.3390/s20164495
Reynaud L, Bouchet-Spinelli A, Raillon C, Buhot A. Sensing with Nanopores and Aptamers: A Way Forward. Sensors. 2020; 20(16):4495. https://doi.org/10.3390/s20164495
Chicago/Turabian StyleReynaud, Lucile, Aurélie Bouchet-Spinelli, Camille Raillon, and Arnaud Buhot. 2020. "Sensing with Nanopores and Aptamers: A Way Forward" Sensors 20, no. 16: 4495. https://doi.org/10.3390/s20164495






