Macro-, Micro- and Nano-Roughness of Carbon-Based Interface with the Living Cells: Towards a Versatile Bio-Sensing Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Substrate Synthesis
2.1.1. Black Silicon (bSi)
2.1.2. PyC on SiO2 and PyC on bSi
2.1.3. Graphene Nanowalls (GNW)
2.2. Structural and Morphological Characterization
2.2.1. Scanning Electron Microscopy (SEM)
2.2.2. Atomic Force Microscopy (AFM)
2.2.3. Raman Spectroscopy
2.3. Cell Culture Growth and Visualization
2.3.1. Cell Culture
2.3.2. Fluorescence Microscopy
2.3.3. Data Analysis
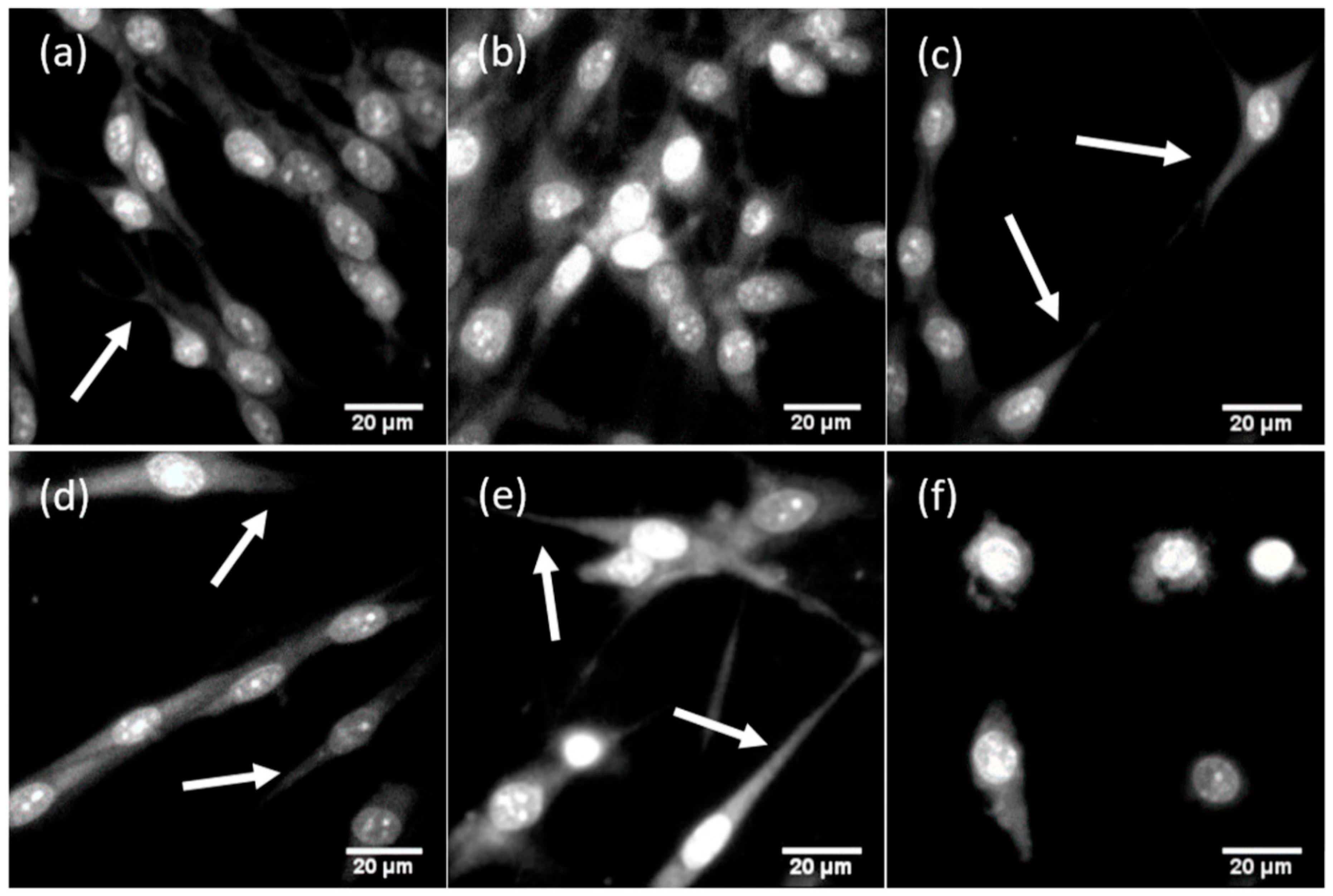
3. Results
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dahinden, C.A.; Fehr, J.; Hugli, T.E. Role of Cell Surface Contact in the Kinetics of Superoxide Production by Granulocytes. J. Clin. Investig. 1983, 72, 113–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spillmann, C.M.; Lomakina, E.; Waugh, R.E. Neutrophil Adhesive Contact Dependence on Impingement Force. Biophys. J. 2004, 87, 4237–4245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, R.; Benz, E.J.; Silberstein, L.E.; Heslop, H.E.; Weitz, J.I.; Anastasi, J.; Salama, M.E.; Abutalib, S. Hematology: Basic Principles and Practice; Elsevier Inc.: Amsterdam, The Netherlands, 2017. [Google Scholar] [CrossRef]
- Chen, S.; Lewallen, M.; Xie, T. Adhesion in the Stem Cell Niche: Biological Roles and Regulation. Development (Cambridge). Development 2013, 140, 255–265. [Google Scholar] [CrossRef] [Green Version]
- Bian, S. Cell Adhesion Molecules in Neural Stem Cell and Stem Cell—Based Therapy for Neural Disorders. In Neural Stem Cells—New Perspectives; InTech: London, UK, 2013. [Google Scholar] [CrossRef] [Green Version]
- Akhmanova, M.; Osidak, E.; Domogatsky, S.; Rodin, S.; Domogatskaya, A. Review Article Physical, Spatial, and Molecular Aspects of Extracellular Matrix of In Vivo Niches and Artificial Scaffolds Relevant to Stem Cells Research. Stem Cells Int. 2015, 2015, 167025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahrami, S.; Baheiraei, N.; Mohseni, M.; Razavi, M.; Ghaderi, A.; Azizi, B.; Rabiee, N.; Karimi, M. Three-Dimensional Graphene Foam as a Conductive Scaffold for Cardiac Tissue Engineering. J. Biomater. Appl. 2019, 34, 74–85. [Google Scholar] [CrossRef]
- Costa, F.; Carvalho, I.F.; Montelaro, R.C.; Gomes, P.; Martins, M.C. Covalent immobilization of antimicrobial peptides (AMPs) onto biomaterial surfaces. Acta Biomater. 2011, 7, 1431–1440. [Google Scholar] [CrossRef] [Green Version]
- Krutty, J.D.; Schmitt, S.K.; Gopalan, P.; Murphy, W.L. Surface functionalization and dynamics of polymeric cell culture substrates. Curr. Opin. Biotechnol. 2016, 40, 164–169. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Li, Y.; Tjong, S.C. Graphene Nanomaterials: Synthesis, Biocompatibility, and Cytotoxicity. Int. J. Mol. Sci. 2018, 19, 3564. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.T.; Choi, M.K.; Sim, Y.; Lim, J.T.; Kim, G.S.; Seong, M.J.; Hyung, J.H.; Kim, K.S.; Umar, A.; Lee, S.K. Effect of Graphene Oxide Ratio on the Cell Adhesion and Growth Behavior on a Graphene Oxide-Coated Silicon Substrate. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Syama, S.; Mohanan, P.V. Comprehensive Application of Graphene: Emphasis on Biomedical Concerns. Nano-Micro Lett. 2019, 11, 1–31. [Google Scholar] [CrossRef] [Green Version]
- Turon Teixidor, G.; Gorkin, R.A.; Tripathi, P.P.; Bisht, G.S.; Kulkarni, M.; Maiti, T.K.; Battacharyya, T.K.; Subramaniam, J.R.; Sharma, A.; Park, B.Y.; et al. Carbon Microelectromechanical Systems as a Substratum for Cell Growth. Biomed. Mater. 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Madhusoodan, A.; Mili, B.; Kumar, A.; Saxena, A.C.; Kumar, K.; Sarkar, M.; Singh, P.; Srivastava, S.; Bag, S. Functionalized Carbon Nanotubes as Suitable Scaffold Materials for Proliferation and Differentiation of Canine Mesenchymal Stem Cells. Int. J. Nanomed. 2017, 12, 3235–3252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, G.Z.; Kim, M.; Shin, U.S.; Kim, H.W. Neurite Outgrowth of Dorsal Root Ganglia Neurons Is Enhanced on Aligned Nanofibrous Biopolymer Scaffold with Carbon Nanotube Coating. Neurosci. Lett. 2011, 501, 10–14. [Google Scholar] [CrossRef]
- Fabbro, A.; Bosi, S.; Ballerini, L.; Prato, M. Carbon Nanotubes: Artificial Nanomaterials to Engineer Single Neurons and Neuronal Networks. ACS Chem. Neurosci. 2012, 3, 611–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bei, H.P.; Yang, Y.; Zhang, Q.; Tian, Y.; Luo, X.; Yang, M.; Zhao, X. Graphene-Based Nanocomposites for Neural Tissue Engineering. Molecules 2019, 24, 658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menaa, F.; Abdelghani, A.; Menaa, B. Graphene Nanomaterials as Biocompatible and Conductive Scaffolds for Stem Cells: Impact for Tissue Engineering and Regenerative Medicine. J. Tissue Eng. Regen. Med. 2015, 9, 1321–1338. [Google Scholar] [CrossRef]
- Ryu, S.; Kim, B.S. Culture of Neural Cells and Stem Cells on Graphene. Tissue Eng. Regen. Med. 2013, 10, 39–46. [Google Scholar] [CrossRef]
- Li, N.; Zhang, Q.; Gao, S.; Song, Q.; Huang, R.; Wang, L.; Liu, L.; Dai, J.; Tang, M.; Cheng, G. Three-Dimensional Graphene Foam as a Biocompatible and Conductive Scaffold for Neural Stem Cells. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [Green Version]
- Mofazzal Jahromi, M.A.; Abdoli, A.; Rahmanian, M.; Bardania, H.; Bayandori, M.; Moosavi Basri, S.M.; Kalbasi, A.; Aref, A.R.; Karimi, M.; Hamblin, M.R. Microfluidic Brain-on-a-Chip: Perspectives for Mimicking Neural System Disorders. Mol. Neurobiol. 2019, 56, 8489–8512. [Google Scholar] [CrossRef]
- Lotti, F.; Ranieri, F.; Vadalà, G.; Zollo, L.; Di Pino, G. Invasive Intraneural Interfaces: Foreign Body Reaction Issues. Frontiers in Neuroscience. Front. Media 2017, 11, 497. [Google Scholar] [CrossRef]
- Kleshch, V.I.; Vasilyeva, E.A.; Lyashenko, S.A.; Obronov, I.V.; Tyurnina, A.V.; Obraztsov, A.N. Surface Structure and Field Emission Properties of Few-Layer Graphene Flakes. Phys. Status Solidi Basic Res. 2011, 248, 2623–2626. [Google Scholar] [CrossRef]
- Kaplas, T.; Svirko, Y. Direct Deposition of Semitransparent Conducting Pyrolytic Carbon Films. J. Nanophotonics 2012, 6, 061703. [Google Scholar] [CrossRef]
- Hiramatsu, M.; Kondo, H.; Hori, M. Graphene Nanowalls. In New Progress on Graphene Research; InTech: London, UK, 2013. [Google Scholar] [CrossRef] [Green Version]
- Batrakov, K.; Kuzhir, P.; Maksimenko, S.; Paddubskaya, A.; Voronovich, S. Enhanced Microwave Shielding Effectiveness of Ultrathin Pyrolytic Carbon Films. Appl. Phys. Lett. 2013, 103, 73117. [Google Scholar] [CrossRef] [Green Version]
- Cook, S.D.; Beckenbaugh, R.D.; Redondo, J.; Popich, L.S.; Klawitter, J.J.; Linscheid, R.L. Long-Term Follow-up of Pyrolytic Carbon Metacarpophalangeal Implants. J. Bone Jt. Surg. Ser. A 1999, 81, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Dovbeshko, G.I.; Romanyuk, V.R.; Pidgirnyi, D.V.; Cherepanov, V.V.; Andreev, E.O.; Levin, V.M.; Kuzhir, P.P.; Kaplas, T.; Svirko, Y.P. Optical Properties of Pyrolytic Carbon Films Versus Graphite and Graphene. Nanoscale Res. Lett. 2015, 10. [Google Scholar] [CrossRef] [Green Version]
- Graham, A.P.; Schindler, G.; Duesberg, G.S.; Lutz, T.; Weber, W. An Investigation of the Electrical Properties of Pyrolytic Carbon in Reduced Dimensions: Vias and Wires. J. Appl. Phys. 2010, 107, 114316. [Google Scholar] [CrossRef]
- McEvoy, N.; Peltekis, N.; Kumar, S.; Rezvani, E.; Nolan, H.; Keeley, G.P.; Blau, W.J.; Duesberg, G.S. Synthesis and Analysis of Thin Conducting Pyrolytic Carbon Films. Carbon 2012, 50, 1216–1226. [Google Scholar] [CrossRef]
- Heikkinen, J.J.; Peltola, E.; Wester, N.; Koskinen, J.; Laurila, T.; Franssila, S.; Jokinen, V. Fabrication of Micro- and Nanopillars from Pyrolytic Carbon and Tetrahedral Amorphous Carbon. Micromachines 2019, 10, 510. [Google Scholar] [CrossRef] [Green Version]
- Shah, A.; Stenberg, P.; Karvonen, L.; Ali, R.; Honkanen, S.; Lipsanen, H.; Peyghambarian, N.; Kuittinen, M.; Svirko, Y.; Kaplas, T. Pyrolytic Carbon Coated Black Silicon. Sci. Rep. 2016, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Ozturk, S.; Kayabasi, E.; Kucukdogan, N.; Ayakdas, O. Progress in Applications of Black Silicon. Most Recent Stud. Sci. Art 2018, 2, 1866–1882. [Google Scholar]
- Sainiemi, L.; Jokinen, V.; Shah, A.; Shpak, M.; Aura, S.; Suvanto, P.; Franssila, S. Non-Reflecting Silicon and Polymer Surfaces by Plasma Etching and Replication. Adv. Mater. 2011, 23, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Hassler, M. Other commonly used biomedical coatings: Pyrolytic carbon coatings. Coat. Biomed. Appl. 2012, 75–105. [Google Scholar] [CrossRef]
- Zhang, W.; Li, A.; Reznik, B.; Deutschmann, O. Thermal expansion of pyrolytic carbon with various textures, ZAMM Zeitschrift Fur Angew. Math. Mech. 2013, 93, 338–345. [Google Scholar] [CrossRef]
- Obraztsov, A.N.; Pavlovsky, I.Y.; Volkov, A.P.; Petrov, A.S.; Petrov, V.I.; Rakova, E.V.; Roddatis, V.V. Electron Field Emission and Structural Properties of Carbon Chemically Vapor-Deposited Films. Diam. Relat. Mater. 1999, 8, 814–819. [Google Scholar] [CrossRef]
- Magdesieva, T.V.; Shvets, P.V.; Nikitin, O.M.; Obraztsova, E.A.; Tuyakova, F.T.; Sergeyev, V.G.; Khokhlov, A.R.; Obraztsov, A.N. Electrochemical characterization of mesoporous nanographite films. Carbon 2016, 105, 96–102. [Google Scholar] [CrossRef]
- Childres, I.; Jauregui, L.A.; Park, W.; Caoa, H.; Chena, Y.P. Raman Spectroscopy of Graphene and Related Materials. In New Developments in Photon and Materials Research; Nova Science Publishers: Hauppauge, NY, USA, 2013; pp. 403–418. [Google Scholar]
- Baryshevsky, V.; Belous, N.; Gurinovich, A.; Gurnevich, E.; Kuzhir, P.; Maksimenko, S.; Molchanov, P.; Shuba, M.; Roddatis, V.; Kaplas, T.; et al. Study of nanometric thin pyrolytic carbon films for explosive, electron emission cathode in high-voltage planar diode. In Thin Solid Films; Elsevier: Amsterdam, The Netherlands, 2015; pp. 107–111. [Google Scholar] [CrossRef]
- Lambin, P. Graphene as a prototypical model for two-dimensional continuous mechanics. Appl. Sci. 2017, 7, 830. [Google Scholar] [CrossRef]
- Samanta, T.; Biswas, R.; Banerjee, S.; Bagchi, B. Study of Distance Dependence of Hydrophobic Force between Two Graphene-like Walls and a Signature of Pressure Induced Structure Formation in the Confined Water. J. Chem. Phys. 2018, 149. [Google Scholar] [CrossRef]
- Riveline, D.; Zamir, E.; Balaban, N.Q.; Schwarz, U.S.; Ishizaki, T.; Narumiya, S.; Kam, Z.; Geiger, B.; Bershadsky, A.D. Focal Contacts as Mechanosensors: Externally Applied Local Mechanical Force Induces Growth of Focal Contacts by an MDia1-Dependent and ROCK-Independent Mechanism. J. Cell Biol. 2001, 153, 1175–1185. [Google Scholar] [CrossRef]
- Owen, G.R.; Meredith, D.O.; Ap Gwynn, I.; Richards, R.G.; Bongrand, P.; Curtis, A.S.G. Focal Adhesion Quantification—A New Assay of Material Biocompatibility? Rev. Eur. Cells Mater. AO Res. Inst. Davos 2005, 85–96. [Google Scholar] [CrossRef]
- Simitzi, C.; Stratakis, E.; Fotakis, C.; Athanassakis, I.; Ranella, A. Microconical Silicon Structures Influence NGF-Induced PC12 Cell Morphology. J. Tissue Eng. Regen. Med. 2015, 9, 424–434. [Google Scholar] [CrossRef]



| Parameter | PyC (20 nm) on SiO2 | PyC (40 nm) on SiO2 | bSi | PyC (20 nm) on bSi |
|---|---|---|---|---|
| Surface Area Difference, % | 0.3 ± 0.2 | 2.9 ± 2.5 | 136 ± 7 | 139 ± 9 |
| Image Rq, nm | 0.9 ± 0.2 | 2.9 ± 1.3 | 81 ± 12 | 75 ± 9 |
| Image Rmax, nm | 11.2 ± 2.6 | 42 ± 19 | 499 ± 12 | 508 ± 37 |
| Mode | PyC (40 nm) on SiO2 | PyC (20 nm) on SiO2 | PyC (20 nm) on bSi | GNWs on Flat Si |
|---|---|---|---|---|
| D mode, cm−1 | 1322.6 ± 2.1 | 1307.2 ± 1.3 | 1322.2 ± 2.2 | 1314.7 ± 3.1 |
| G mode, cm−1 | 1595.2 ± 1.8 | 1602.1 ± 3.4 | 1595.2 ± 2.6 | 1580.8 ± 4.7 |
| Parameter | Plastic | PyC (20 nm) on SiO2 | PyC (40 nm) on SiO2 | bSi | PyC (20 nm) on bSi | Graphene Nanowalls |
|---|---|---|---|---|---|---|
| Area occupied by cells, % | 63.3 ± 10.3 | 33.7 ± 7.1 * | 55.1 ± 10.3 | 68.9 ± 28.2 | # 20.7 ± 4.6 * | 11.6 ± 5.1 * |
| Area occupied by cells, µm2 | 25310 ± 4131 | 13475 ± 2848 * | 22052 ± 4124 | 27570 ± 11261 | # 8271 ± 1855 * | 4652 ± 2047 * |
| Average cell number per 200 × 200 µm area | 36.0 ± 3.7 | 15.6 ± 1.1 * | 25.2 ± 2.6 * | 53.8 ± 15.2 *** | ## 13.0 ± 0.7 * | 7.3 ± 0.4 * |
| Area per cell, % | 1.8 ± 0.3 | 2.2 ± 0.1 ** | 2.2 ± 0.1 ** | 1.3 ± 0.3 *** | 1.6 ± 0.1 | 1.6 ± 0.1 |
| Area per cell, µm2 | 703 ± 115 | 864 ± 183 ** | 875 ± 164 ** | 513 ± 209 *** | 636 ± 143 | 637 ± 280 |
| Cell morphology characteristics | Elongated, spindle-shaped, 2–3 long processes, numerous intercellular contacts | Elongated, spindle-shaped, 2–3 long thin processes, few intercellular contacts | Elongated, spindle-shaped, 2–3 long thin processes, few intercellular contacts | Spindle-shaped, rounded, many middle-sized processes, many intercellular contacts | Elongated, spindle-shaped, 2–3 long thin processes, few intercellular contacts | Rounded, no processes or short wide processes, no intercellular contacts |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golubewa, L.; Rehman, H.; Kulahava, T.; Karpicz, R.; Baah, M.; Kaplas, T.; Shah, A.; Malykhin, S.; Obraztsov, A.; Rutkauskas, D.; et al. Macro-, Micro- and Nano-Roughness of Carbon-Based Interface with the Living Cells: Towards a Versatile Bio-Sensing Platform. Sensors 2020, 20, 5028. https://doi.org/10.3390/s20185028
Golubewa L, Rehman H, Kulahava T, Karpicz R, Baah M, Kaplas T, Shah A, Malykhin S, Obraztsov A, Rutkauskas D, et al. Macro-, Micro- and Nano-Roughness of Carbon-Based Interface with the Living Cells: Towards a Versatile Bio-Sensing Platform. Sensors. 2020; 20(18):5028. https://doi.org/10.3390/s20185028
Chicago/Turabian StyleGolubewa, Lena, Hamza Rehman, Tatsiana Kulahava, Renata Karpicz, Marian Baah, Tommy Kaplas, Ali Shah, Sergei Malykhin, Alexander Obraztsov, Danielis Rutkauskas, and et al. 2020. "Macro-, Micro- and Nano-Roughness of Carbon-Based Interface with the Living Cells: Towards a Versatile Bio-Sensing Platform" Sensors 20, no. 18: 5028. https://doi.org/10.3390/s20185028








