Bacterial Respiration Used as a Proxy to Evaluate the Bacterial Load in Cooling Towers
Abstract
:1. Introduction
2. Materials and Methods
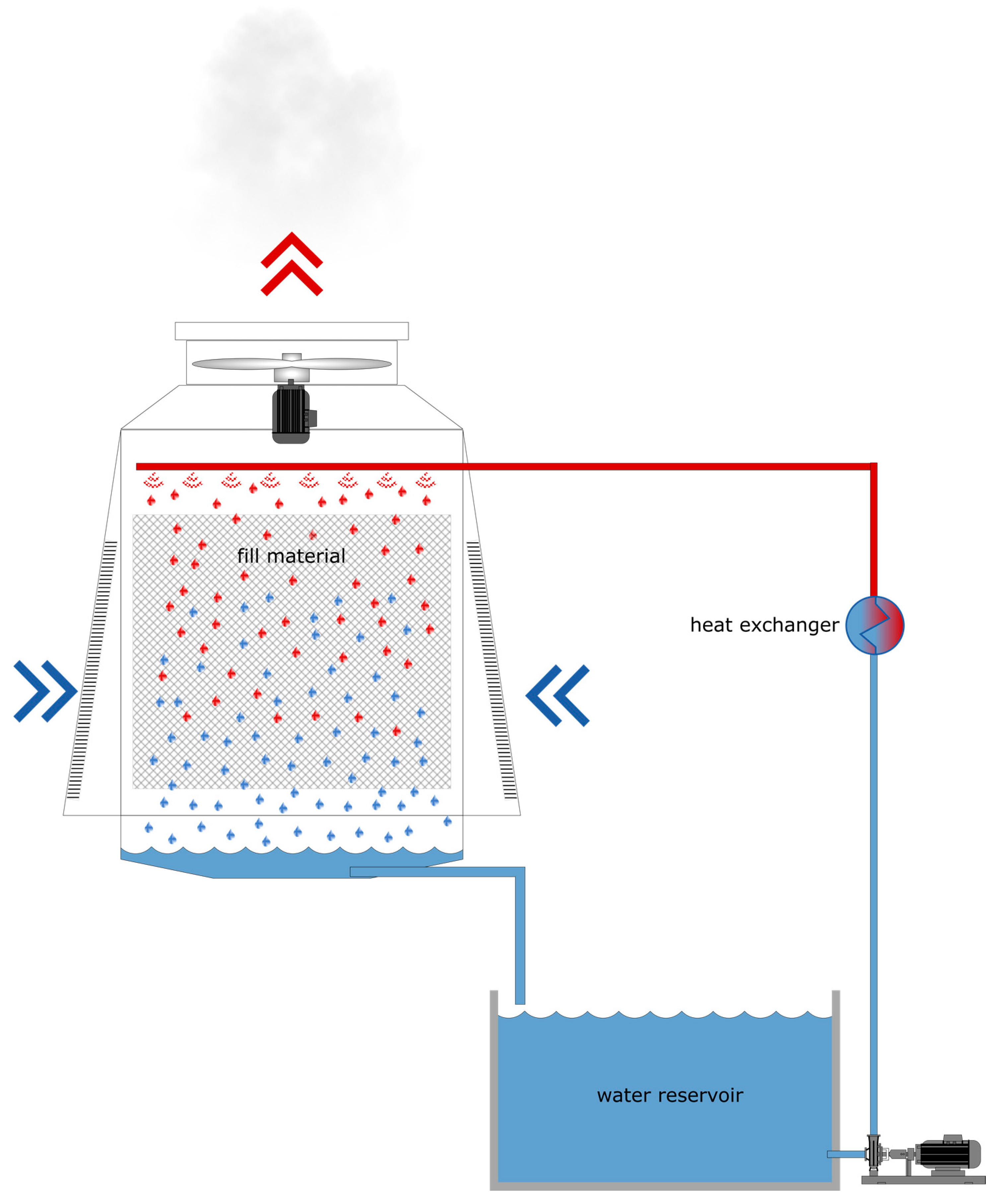
2.1. Sampling
2.2. Quantitative Analyses
2.2.1. Cultivation-Based Methods
2.2.2. Flow Cytometry
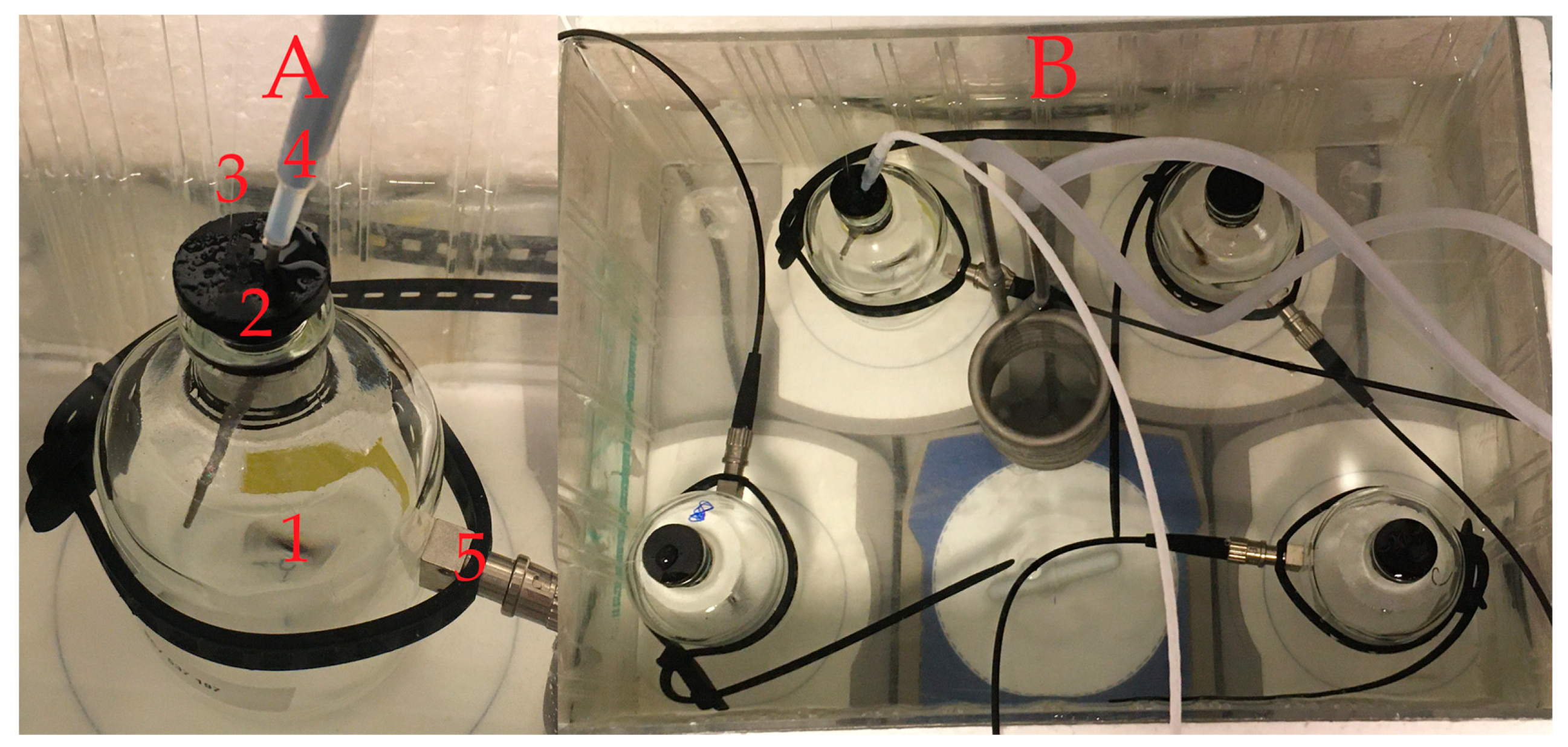
2.2.3. Oxygen Respiration Measurements
2.2.4. Bacterial Community Analysis
3. Results and Discussion
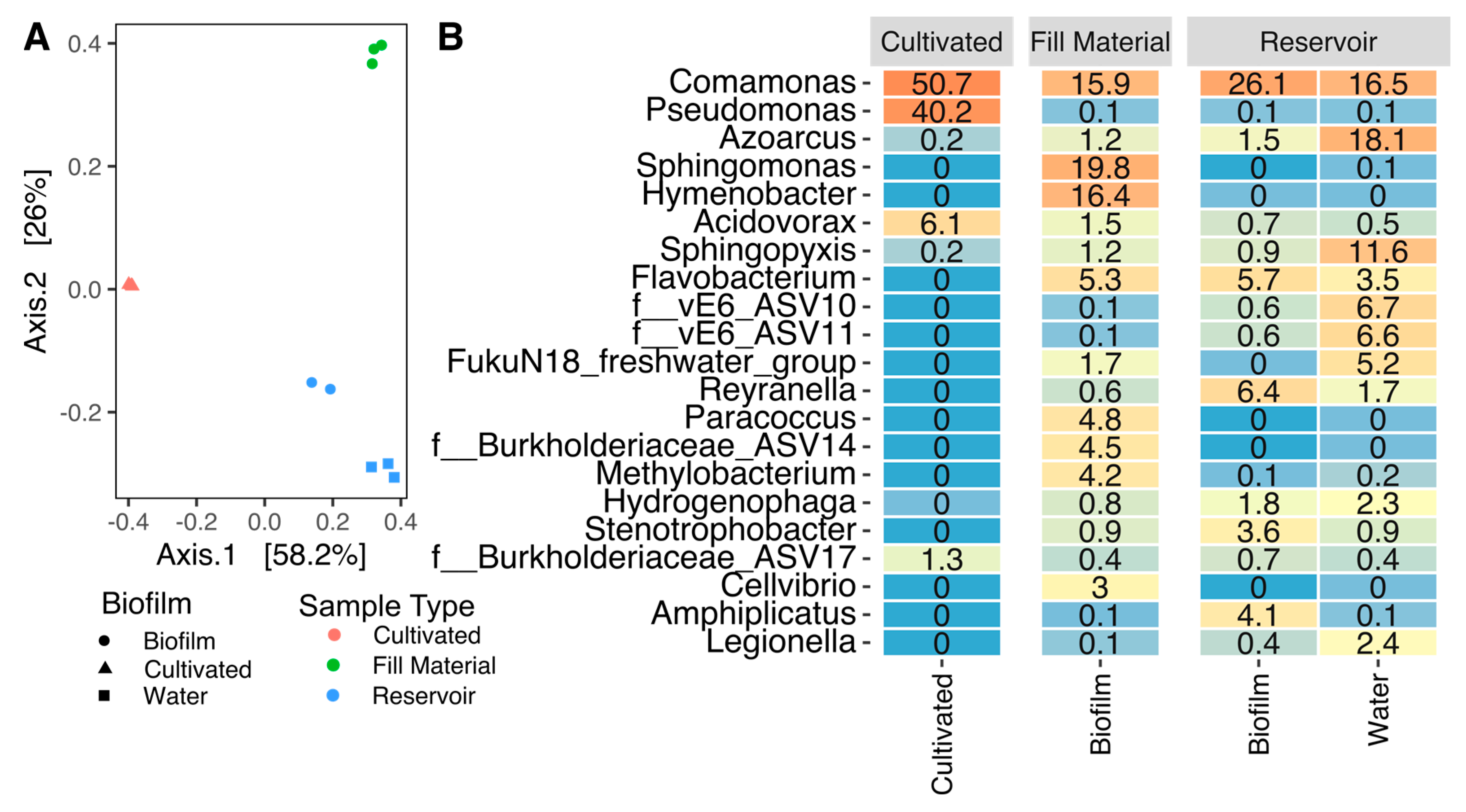
3.1. Cultivation-Based Methods and Community Analysis
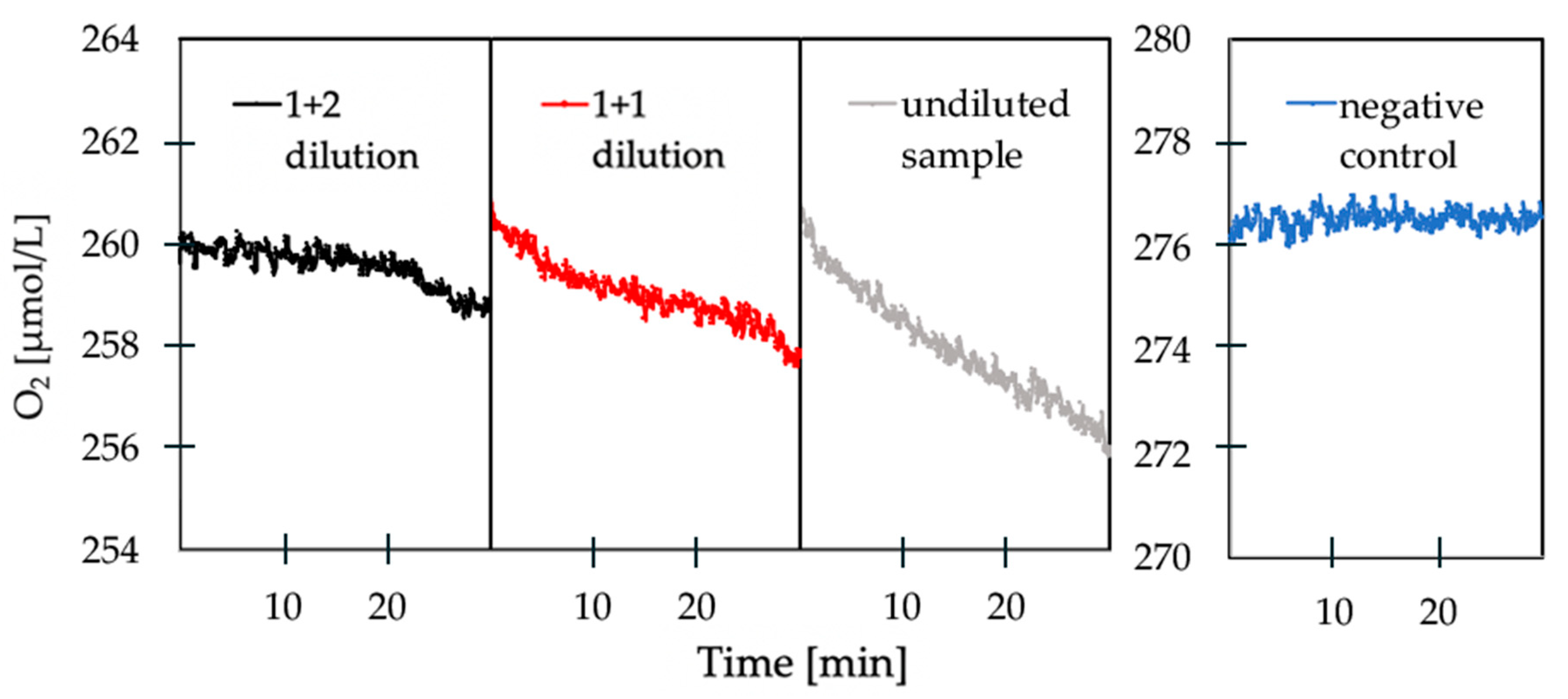
3.2. Oxygen Respiration Measurements
4. Conclusions and Perspective
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Altman, S.J.; Jensen, R.P. Membrane treatment of side-stream cooling tower water for reduction of water usage. Desalination 2012, 285, 177–183. [Google Scholar] [CrossRef]
- Wagner, T.V.; Parsons, J.R. A review on the removal of conditioning chemicals from cooling tower water in constructed wetlands. Crit. Rev. Environ. Sci. Technol. 2018, 48, 1094–1125. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, W. Role of bacterial adhesion in the microbial ecology of biofilms in cooling tower systems. Biofouling 2009, 25, 241–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.C. Biofouling and me: My Stockholm syndrome with biofilms. Water Res. 2020, 173, 115576. [Google Scholar] [CrossRef]
- Muller, H. Fouling of Heat Exchangers Surfaces; Springer: Berlin/Heidelberg, Germany, 2010; pp. 79–104. [Google Scholar]
- Chandrasatheesh, C.; Jayapriya, J. Biocorrosion. In Bioelectrochemical Interface Engineering; John Wiley & Sons, Inc.: 111 River Street, Hoboken, NJ, USA, 2019; pp. 77–90. [Google Scholar]
- Narenkumar, J.; Alsalhi, M.S.; Arul Prakash, A.; Abilaji, S.; Devanesan, S.; Rajasekar, A.; Alfuraydi, A.A. Impact and Role of Bacterial Communities on Biocorrosion of Metals Used in the Processing Industry. Acs Omega 2019, 4, 21353–21360. [Google Scholar] [CrossRef] [Green Version]
- Meesters, K.P.H.; Van Groenestijn, J.W. Biofouling reduction in recirculating cooling systems through biofiltration of process water. Water Res. 2003, 37, 525–532. [Google Scholar] [CrossRef]
- Arnow, P.M.; Chou, T. Nosocomial legionnaires’ disease caused by aerosolized tap water from respiratory devices. J. Infect. Dis. 1982, 146, 460–467. [Google Scholar] [CrossRef]
- Tsao, H.-F.; Scheikl, U.; Herbold, C.; Indra, A.; Walochnik, J.; Horn, M. The cooling tower water microbiota: Seasonal dynamics and co-occurrence of bacterial and protist phylotypes. Water Res. 2019, 159, 464–479. [Google Scholar] [CrossRef]
- Engel, G.L. Isolation of a Bacterium and Demonstration of Its Role in Other Respiratory Disease. N. Engl. J. Med. 1977, 297, 1197–1203. [Google Scholar]
- Špaleková, M.; Kotrbancová, M.; Fulová, M.; Šimonyiová, D. Risk of Legionellosis from Exposure to Water Aerosol from Industrial Cooling Tower. Int. Arch. Public Heal. Community Med. 2019, 3, 1–8. [Google Scholar] [CrossRef]
- Fliermans, C.B.; Cherry, W.B. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 1981, 41, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Fraser, D.W.; Tsai, T.R. Legionnaires’ Disease: Description of an Epidemic of Pneumonia. N. Engl. J. Med. 1977, 297, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Dondero, T.; Rendtorff, R. An outbreak of Legionnaires’ disease associated with a contaminated air-conditioning cooling tower. Chin. Med. J. (Engl). 1980, 302, 365–370. [Google Scholar] [CrossRef] [PubMed]
- García-fulgueiras, A.; Navarro, C. Legionella Outbreak in Spain, 2001. Emerg. Infect. Dis. 2003, 9, 915–921. [Google Scholar] [CrossRef]
- Pereira, R.P.A.; Peplies, J. Bacterial community dynamics in a cooling tower with emphasis on pathogenic bacteria and Legionella species using universal and genus-specific deep sequencing. Water Res. 2017, 122, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Nhu Nguyen, T.M.; Ilef, D.; Jarraud, S.; Rouil, L.; Campese, C.; Che, D.; Haeghebaert, S.; Ganiayre, F.; Marcel, F.; Etienne, J.; et al. A Community--Wide Outbreak of Legionnaires Disease Linked to Industrial Cooling Towers—How Far Can Contaminated Aerosols Spread? J. Infect. Dis. 2006, 193, 102–111. [Google Scholar] [CrossRef] [Green Version]
- Walser, S.M.; Gerstner, D.G.; Brenner, B.; Höller, C.; Liebl, B.; Herr, C.E.W. Assessing the environmental health relevance of cooling towers – A systematic review of legionellosis outbreaks. Int. J. Hyg. Env. Health 2014, 217, 145–154. [Google Scholar] [CrossRef]
- Lee, S. An Overview of the European Technical Guidelines for the Prevention, Control and Investigation of Infections Caused by Legionella species. Perspect. Public Health 2018, 138, 241–247. [Google Scholar] [CrossRef]
- Donachie, S.P.; Foster, J.S.; Brown, M.V. Culture clash: Challenging the dogma of microbial diversity. Isme J. 2007, 1, 97–99. [Google Scholar] [CrossRef]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. Isme J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet.J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, K.; Kirkegaard, R.; Karst, S.; Albertsen, M. ampvis2: An R package to analyse and visualise 16S rRNA amplicon data. bioRxiv 2018. [Google Scholar]
- Pinel, I.S.M.; Moed, D.H.; Vrouwenvelder, J.S.; van Loosdrecht, M.C.M. Bacterial community dynamics and disinfection impact in cooling water systems. Water Res. 2020, 172, 115505. [Google Scholar] [CrossRef]
- Staley, J.T.; Konopka, A. Measurement of in Situ Activities of Nonphotosynthetic Microorganisms in Aquatic and Terrestrial Habitats. Annu. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef]
- Lloyd, K.G.; Steen, A.D.; Ladau, J.; Yin, J.; Crosby, L. Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes. mSystems 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Paniagua, A.T.; Paranjape, K.; Hu, M.; Bédard, E.; Faucher, S.P. Impact of temperature on Legionella pneumophila, its protozoan host cells, and the microbial diversity of the biofilm community of a pilot cooling tower. Sci. Total Env. 2020, 712. [Google Scholar] [CrossRef]
- Elisseeva, S.; Kelly, C.; Cruz-Romero, M.; Zhdanov, A.V.; Kerry, J.P.; Papkovsky, D.B. The use of optical oxygen sensing and respirometry to quantify the effects of antimicrobials on common food spoilage bacteria and food samples. Sens. ActuatorsB Chem. 2020, 322, 128572. [Google Scholar] [CrossRef]
- Jasionek, G.; Ogurtsov, V.; Papkovsky, D. Rapid detection and respirometric profiling of aerobic bacteria on panels of selective media. J. Appl. Microbiol. 2013, 114, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Robledo, E.; Padilla, C.C.; Aldunate, M.; Stewart, F.J.; Ulloa, O.; Paulmier, A.; Gregori, G.; Revsbech, N.P. Cryptic oxygen cycling in anoxic marine zones. Proc. Natl. Acad. Sci. USA 2017, 114, 8319–8324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koren, K.; Kühl, M. CHAPTER 7. Optical O2 Sensing in Aquatic Systems and Organisms. In Quenched-phosphorescence Detection of Molecular Oxygen: Applications in Life Sciences; Papkovsky, D.B., Dmitriev, R.I., Eds.; Royal Society of Chemistry: Cambridge, UK, 2018; Volume 1, pp. 145–174. [Google Scholar]
- Tahirbegi, I.B.; Ehgartner, J.; Sulzer, P.; Zieger, S.; Kasjanow, A.; Paradiso, M.; Strobl, M.; Bouwes, D.; Mayr, T. Fast pesticide detection inside microfluidic device with integrated optical pH, oxygen sensors and algal fluorescence. Biosens. Bioelectron. 2017, 88, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhao, C.; Xia, L.; Yang, F.; Chang, X.; Wang, Y. Biofouling characteristics and identification of preponderant bacteria at different nutrient levels in batch tests of a recirculating cooling water system. Environ. Technol. 2011, 32, 901–910. [Google Scholar] [CrossRef]
- Riedel, T.E.; Berelson, W.M.; Nealson, K.H.; Finkel, S.E. Oxygen Consumption Rates of Bacteria under Nutrient-Limited Conditions. Appl. Environ. Microbiol. 2013, 79, 4921–4931. [Google Scholar] [CrossRef] [Green Version]
- Lehner, P.; Larndorfer, C.; Garcia-Robledo, E.; Larsen, M.; Borisov, S.M.; Revsbech, N.-P.; Glud, R.N.; Canfield, D.E.; Klimant, I. LUMOS - A Sensitive and Reliable Optode System for Measuring Dissolved Oxygen in the Nanomolar Range. PLoS ONE 2015, 10, e0128125. [Google Scholar] [CrossRef] [Green Version]
- Koren, K.; Borisov, S.M.; Klimant, I. Stable optical oxygen sensing materials based on click-coupling of fluorinated platinum(II) and palladium(II) porphyrins—A convenient way to eliminate dye migration and leaching. Sens. Actuators B Chem. 2012, 169, 173–181. [Google Scholar] [CrossRef] [Green Version]
- Koren, K.; Hutter, L.; Enko, B.; Pein, A.; Borisov, S.M.; Klimant, I. Tuning the dynamic range and sensitivity of optical oxygen-sensors by employing differently substituted polystyrene-derivatives. Sens. Actuators B Chem. 2013. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toman, S.; Kiilerich, B.; Marshall, I.P.G.; Koren, K. Bacterial Respiration Used as a Proxy to Evaluate the Bacterial Load in Cooling Towers. Sensors 2020, 20, 6398. https://doi.org/10.3390/s20216398
Toman S, Kiilerich B, Marshall IPG, Koren K. Bacterial Respiration Used as a Proxy to Evaluate the Bacterial Load in Cooling Towers. Sensors. 2020; 20(21):6398. https://doi.org/10.3390/s20216398
Chicago/Turabian StyleToman, Stepan, Bruno Kiilerich, Ian P.G. Marshall, and Klaus Koren. 2020. "Bacterial Respiration Used as a Proxy to Evaluate the Bacterial Load in Cooling Towers" Sensors 20, no. 21: 6398. https://doi.org/10.3390/s20216398
APA StyleToman, S., Kiilerich, B., Marshall, I. P. G., & Koren, K. (2020). Bacterial Respiration Used as a Proxy to Evaluate the Bacterial Load in Cooling Towers. Sensors, 20(21), 6398. https://doi.org/10.3390/s20216398




