Classification of Watermelon Seeds Using Morphological Patterns of X-ray Imaging: A Comparison of Conventional Machine Learning and Deep Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Image Acquisition
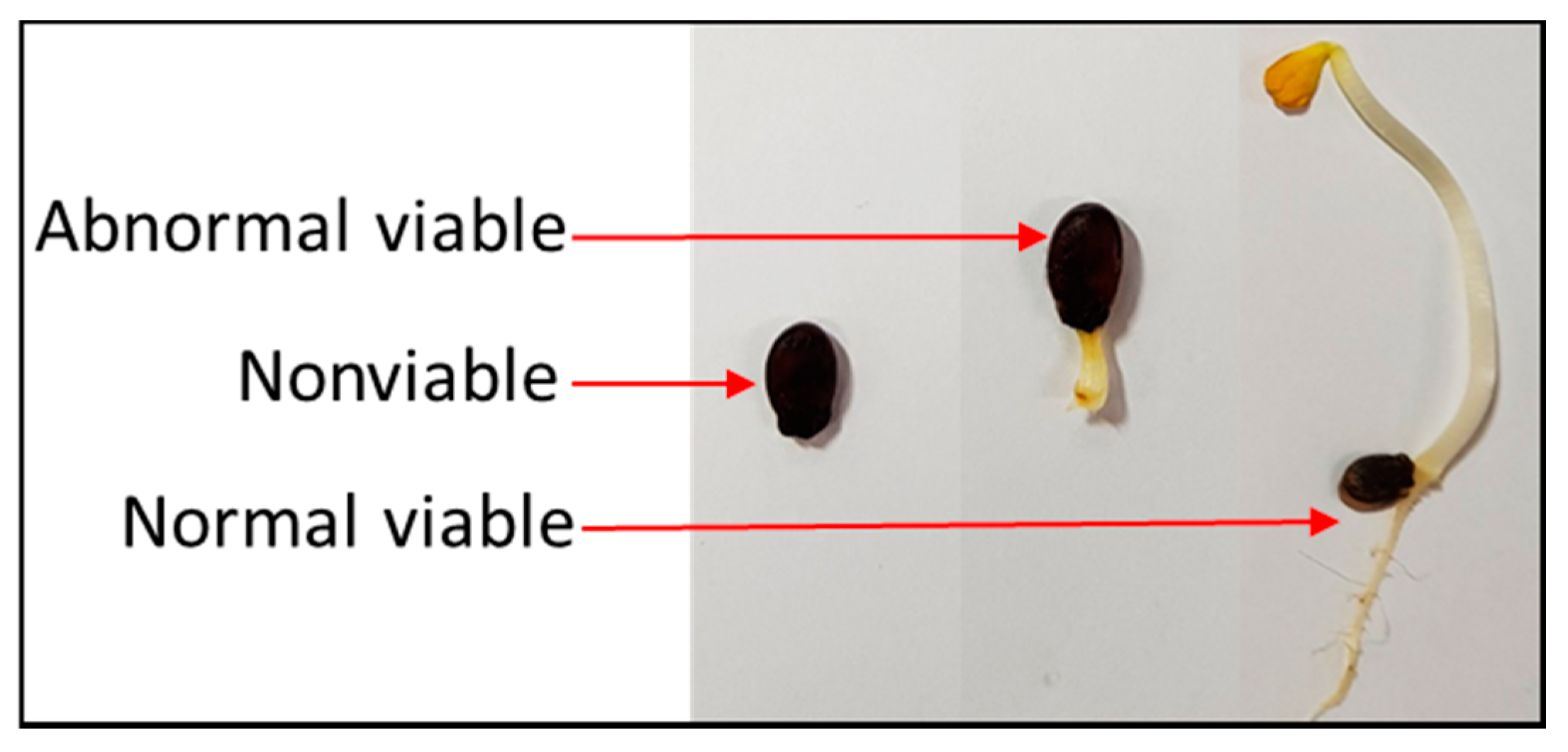
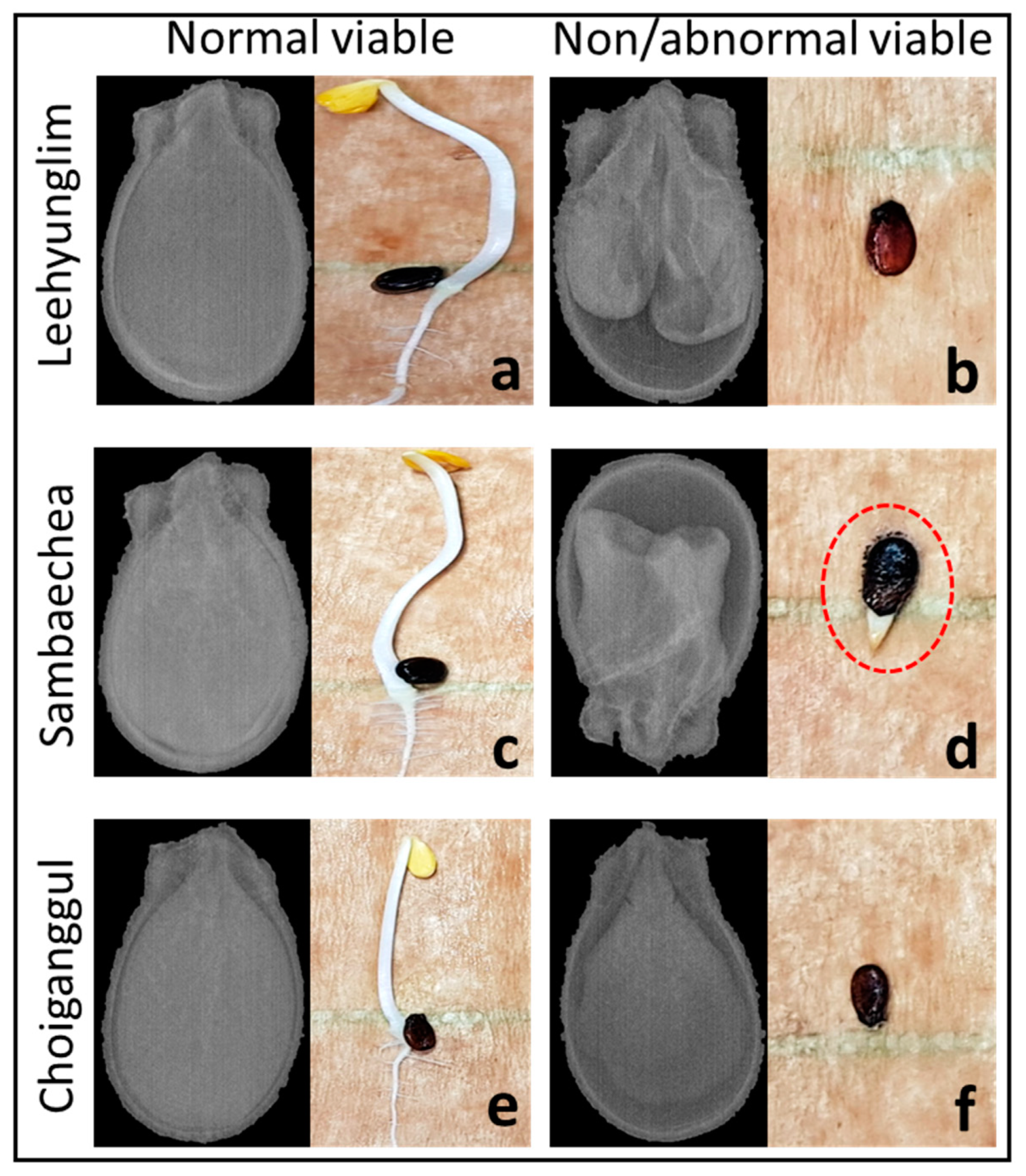
2.3. Germination Test
2.4. Image Preprocessing
2.5. Image Cropping
2.6. Image Masking and Quality Enhancement
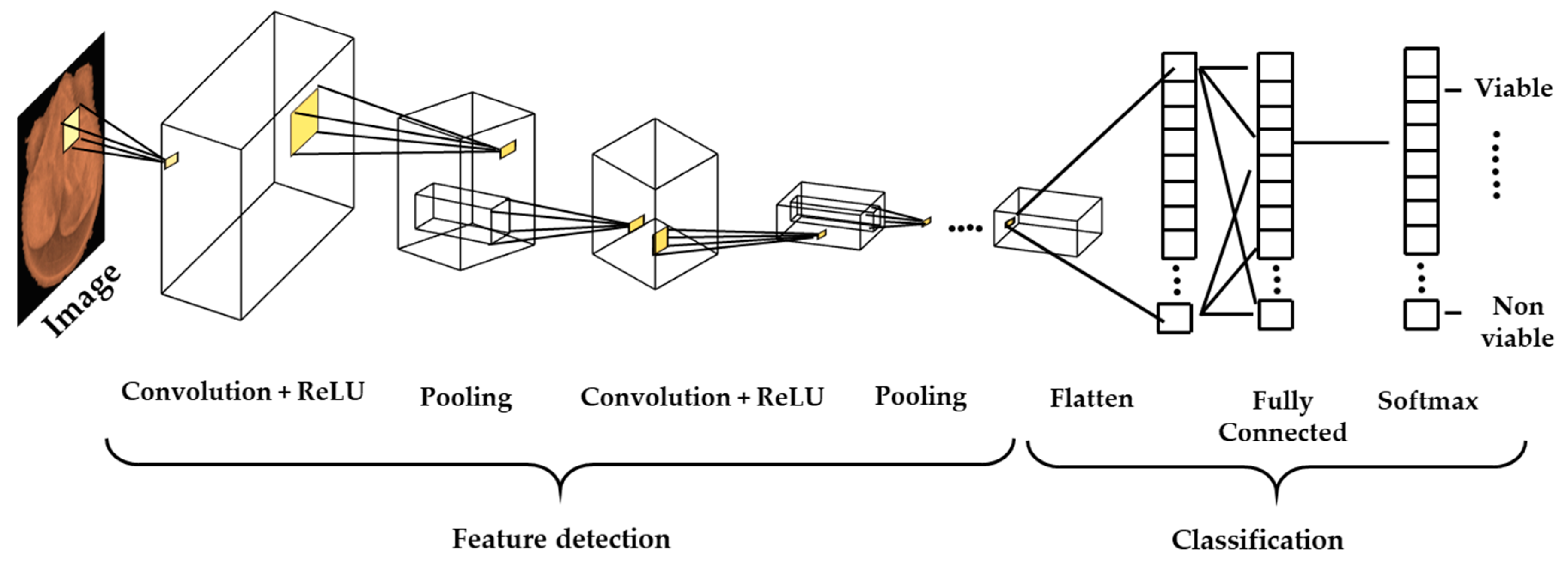
2.7. Feature Extraction, Selection, and Classification
2.8. Transfer Learning
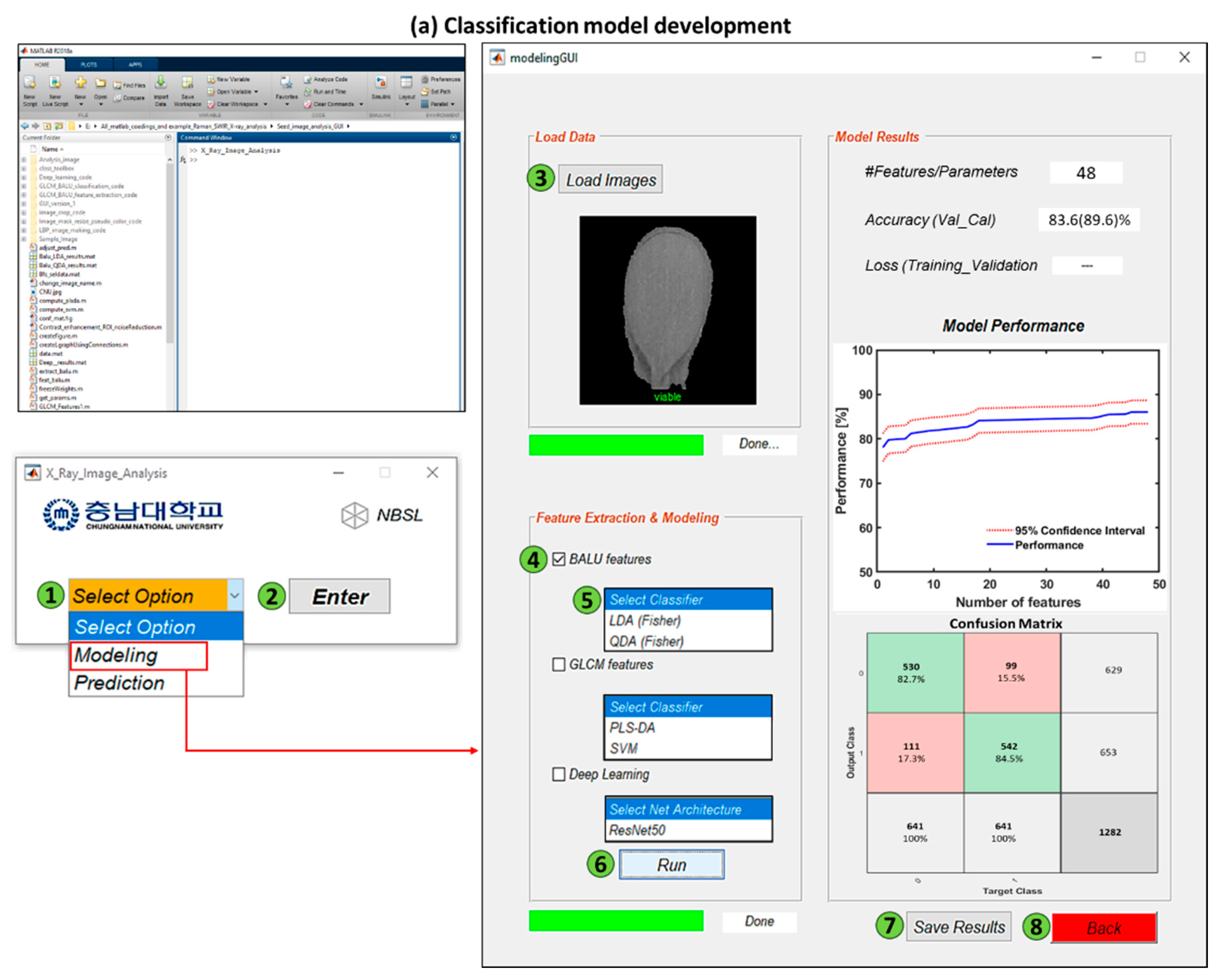
2.9. Analysis Software
3. Results and Discussion
3.1. Germination Test Result
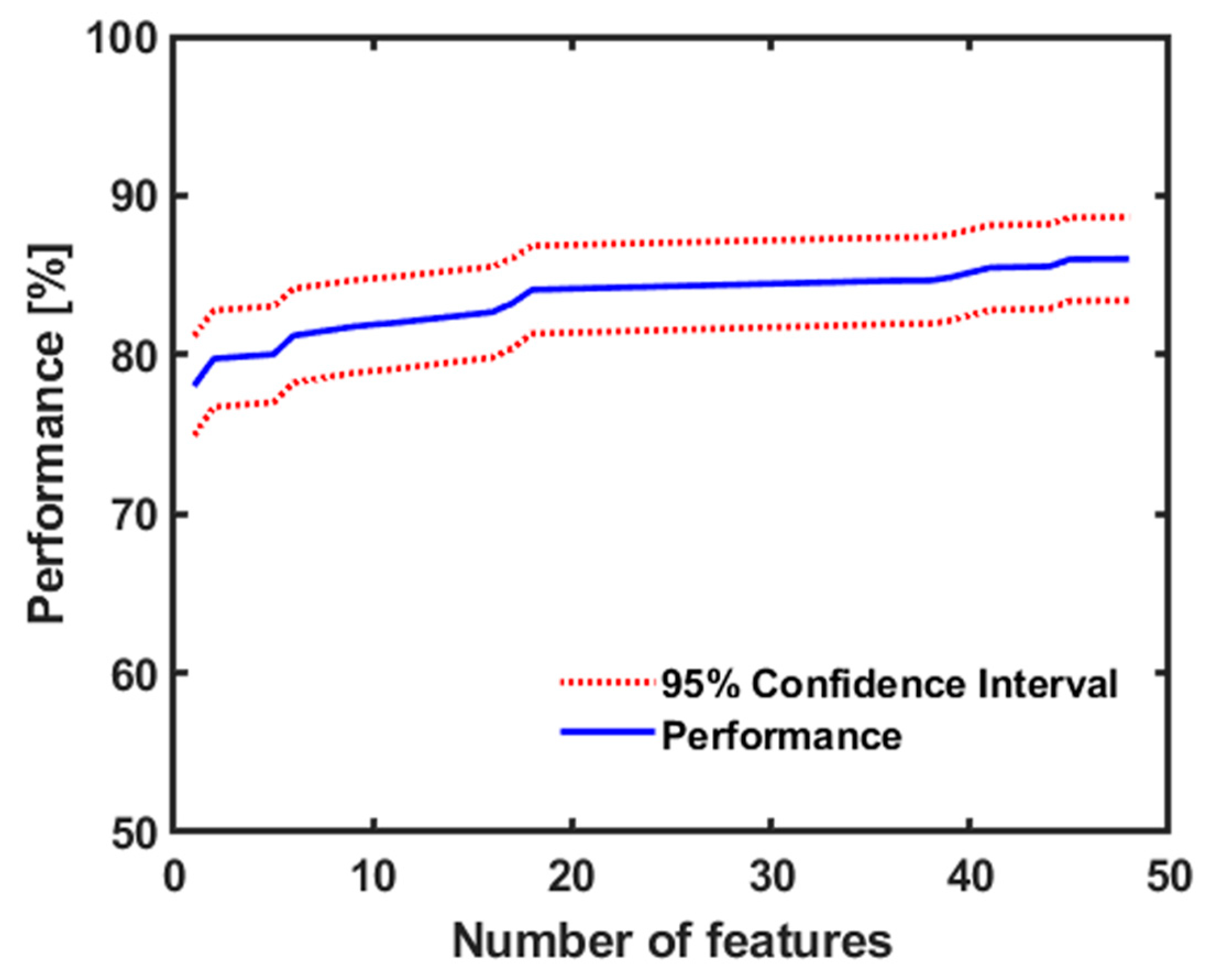
3.2. Conventional Machine Learning
3.3. Transfer Learning
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bewley, J.D.; Black, M. Seeds. In Seeds; Springer: Boston, MA, USA, 1994; pp. 1–33. [Google Scholar]
- Abud, H.F.; Cicero, S.M.; Gomes-Junior, F.G. Radiographic images and relationship of the internal morphology and physiological potential of broccoli seeds. Acta Sci. Agron. 2018, 40. [Google Scholar] [CrossRef]
- Medeiros, A.D.D.; Silva, L.J.D.; Pereira, M.D.; Oliveira, A.M.S.; Dias, D.C.F.S. High-throughput phenotyping of brachiaria grass seeds using free access tool for analyzing X-ray images. Ann. Acad. Bras. Cienc. 2020, 92, e20190209. [Google Scholar] [CrossRef] [PubMed]
- Association of Analytical Chemists (AOAC). Official Methods of Analysis, 13th ed.; The Association of Official Analytical Chemists: Arlington, VA, USA, 1980. [Google Scholar]
- Ogawa, Y.; Kondo, N.; Shibusawa, S. Inside quality evaluation of fruit by X-ray image. In Proceedings of the IEEE/ASME International Conference on Advanced Intelligent Mechatronics, AIM, Kobe, Japan, 20–24 July 2003; Institute of Electrical and Electronics Engineers Inc.: Kobe, Japan, 2003; Volume 2, pp. 1360–1365. [Google Scholar]
- Keagy, P.M.; Schatzki, T.F. Machine recognition of weevil damage in wheat radiographs. In Optics in Agriculture and Forestry, Proceedings of the Applications in Optical Science and Engineering, Boston, MA, USA, 16 November 1992; DeShazer, J.A., Meyer, G.E., Eds.; SPIE: Bellingham, WA, USA, 1993; Volume 1836, pp. 108–119. [Google Scholar]
- Karunakaran, C.; Jayas, D.S.; White, N.D.G. Soft X-ray inspection of wheat kernels infested by Sitophilus oryzae. Trans. ASAE 2003, 46, 739. [Google Scholar] [CrossRef]
- Haff, R.P.; Toyofuku, N. X-ray detection of defects and contaminants in the food industry. Sens. Instrum. Food Qual. Saf. 2008, 2, 262–273. [Google Scholar] [CrossRef]
- Jorge, M.H.A.; Ray, D.T. Germination characterization of guayule seed by morphology, mass and, X-ray analysis. Ind. Crops Prod. 2005, 22, 59–63. [Google Scholar] [CrossRef]
- Paradelo, K.; Martins, J.; Soares, I.; De Carvalho, R. X-ray test to evaluate the physiological potencial of Platypodium elegans Vog. Seeds (Fabaceae). Sci. Agropecu. 2016, 7, 305–311. [Google Scholar] [CrossRef][Green Version]
- Sood, S.; Mahajan, S.; Doegar, A.; Das, A. Internal crack detection in kidney bean seeds using X-ray imaging technique. In Proceedings of the International Conference on Advances in Computing, Communications and Informatics (ICACCI), Jaipur, India, 21–24 September 2016; Institute of Electrical and Electronics Engineers Inc.: Jaipur, India, 2016; pp. 2258–2261. [Google Scholar]
- Al-Turki, T.A.; Baskin, C.C. Determination of seed viability of eight wild Saudi Arabian species by germination and X-ray tests. Saudi J. Biol. Sci. 2017, 24, 822–829. [Google Scholar] [CrossRef][Green Version]
- Van der Burg, W.J.; Aartse, J.W.; Van Zwol, R.A.; Jalink, H.; Bino, R.J. Predicting tomato seedling morphology by X-ray analysis of seeds. J. Am. Soc. Hortic. Sci. 1994, 119, 258–263. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; Association for Computing Machinery: New York, NY, USA, 2012; Volume 2, pp. 1097–1105. [Google Scholar]
- Cireşan, D.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Mitosis detection in breast cancer histology images with deep neural networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2013; pp. 411–418. [Google Scholar]
- Taigman, Y.; Yang, M.; Ranzato, M.; Wolf, L. DeepFace: Closing the gap to human-level performance in face verification. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; IEEE Computer Society: Columbus, OH, USA, 2014; pp. 1701–1708. [Google Scholar]
- Grinblat, G.L.; Uzal, L.C.; Larese, M.G.; Granitto, P.M. Deep learning for plant identification using vein morphological patterns. Comput. Electron. Agric. 2016, 127, 418–424. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.; Wang, G.; Zhang, H. Deep learning for plant identification in natural environment. Comput. Intell. Neurosci. 2017, 2017, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.K.; Ijsselmuiden, J.; Hofstee, J.W.; van Henten, E.J. Transfer learning for the classification of sugar beet and volunteer potato under field conditions. Biosyst. Eng. 2018, 174, 50–65. [Google Scholar] [CrossRef]
- Zhu, S.; Zhou, L.; Gao, P.; Bao, Y.; He, Y.; Feng, L. Near-infrared hyperspectral imaging combined with deep learning to identify cotton seed varieties. Molecules 2019, 24, 3268. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Chen, J.; Zhao, Y.; Zhu, S.; He, Y.; Zhang, C. Variety identification of single rice seed using hyperspectral imaging combined with convolutional neural network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef]
- Craviotto, R.M.; Arango, M.R.; Salinas, A.R.; Gibbons, R.; Bergmann, R.; Montero, M.S. A device for automated digital X-ray imaging for seed analysis. Seed Sci. Technol. 2004, 32, 867–871. [Google Scholar] [CrossRef]
- Dos Anjos, A.; Shahbazkia, H.R. Bi-level image thresholding—A fast method. In Proceedings of the First International Conference on Biomedical Electronics and Devices, BIOSIGNALS 2008, Funchal, Madeira, Portugal, 28–31 January 2008; pp. 70–76. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man. Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Ahmed, M.R.; Yasmin, J.; Collins, W.; Cho, B.-K. X-ray CT image analysis for morphology of muskmelon seed in relation to germination. Biosyst. Eng. 2018, 175, 183–193. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D.D. A survey of transfer learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Learning and transferring mid-level image representations using convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1717–1724. [Google Scholar]
- Sun, Y.; Wang, X.; Tang, X. Deep learning face representation from predicting 10,000 classes. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1891–1898. [Google Scholar]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the International Conference on Learning Representations (ICLR 2015), San Diego, CA, USA, 7–9 May 2015; ICLR: San Diego, CA, USA; pp. 1–14. [Google Scholar]
- Hasan, M.; Kotov, A.; Carcone, A.I.; Dong, M.; Naar, S.; Hartlieb, K.B. A study of the effectiveness of machine learning methods for classification of clinical interview fragments into a large number of categories. J. Biomed. Inform. 2016, 62, 21–31. [Google Scholar] [CrossRef]
- Malik, C.P. Seed Deterioration: A Review. Int. J. Life Sci. Biotechnol. Pharma Res. 2013, 2, 374–385. [Google Scholar]
- Sa, I.; Ge, Z.; Dayoub, F.; Upcroft, B.; Perez, T.; McCool, C. DeepFruits: A fruit detection system using deep neural networks. Sensors 2016, 16, 1222. [Google Scholar] [CrossRef] [PubMed]
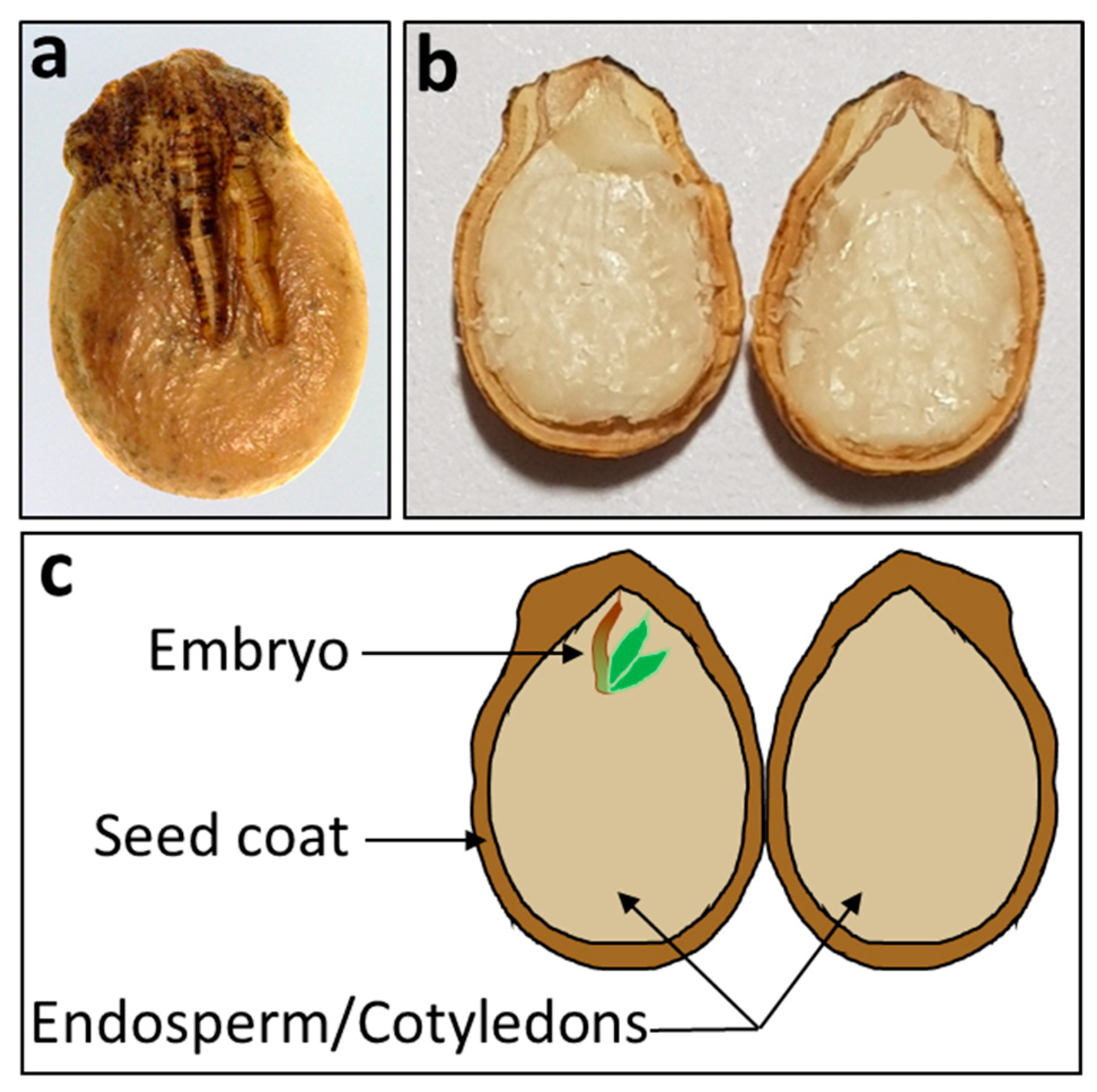



| Parameters | Values |
|---|---|
| System | Xeye-5100F |
| Source voltage | 50 kV |
| Source current | 100 μA |
| Exposure time | 0.05 s |
| Magnification | 18× |
| Filter | Glass effect |
| Cultivars | Viable Seed | Nonviable Seed | Total | Germination Rate |
|---|---|---|---|---|
| Leehyunglim | 72 | 528 | 600 | 12% |
| Sambaechea | 116 | 484 | 600 | 19% |
| Choiganggul | 453 | 147 | 600 | 76% |
| Overall | 641 | 1159 | 1800 | 36% |
| Region of Interest (ROI) | Classifier | 2-Classes Performance (%) | ||
|---|---|---|---|---|
| Mean | UCI a | LCI b | ||
| Whole seed | LDA | 83.6 | 86.1 | 81.1 |
| QDA | 80.8 | 84.4 | 77.2 | |
| KNN (K = 5) | 63.7 | 72.6 | 54.9 | |
| Number | Features Name | Number | Features Names |
|---|---|---|---|
| 1 | i-Gabor(1,1)[Max-A] | 25 | i-LBP(2,2)[8,u2][sd-C] |
| 2 | i-LBP(3,34)[8,u2][sd-C] | 26 | i-LBP(2,26)[8,u2][sd-A] |
| 3 | i-LBP(2,53)[8,u2][sd-B] | 27 | i-LBP(2,29)[8,u2][Max-B] |
| 4 | Fourier Ang (2,1)[rad][Max-C] | 28 | i-LBP(1,36)[8,u2][sd-C] |
| 5 | i-LBP(3,44)[8,u2][sd-A] | 29 | i-LBP(4,42)[8,u2][sd-A] |
| 6 | Fourier Abs (1,1)[Max-C] | 30 | i-LBP(3,51)[8,u2][sd-C] |
| 7 | i-Gabor-J[sd-A] | 31 | i-LBP(2,6)[8,u2][sd-A] |
| 8 | i-LBP(3,27)[8,u2][sd-A] | 32 | i-Intensity Skewness[Max-C] |
| 9 | i-LBP(3,58)[8,u2][Max-A] | 33 | i-LBP(3,2)[8,u2][sd-C] |
| 10 | i-LBP(1,10)[8,u2][Max-C] | 34 | i-LBP(3,41)[8,u2][Max-C] |
| 11 | i-LBP(4,57)[8,u2][sd-C] | 35 | i-LBP(1,57)[8,u2][Max-A] |
| 12 | i-LBP(3,56)[8,u2][Max-C] | 36 | i-LBP(1,5)[8,u2][Max-A] |
| 13 | i-LBP(1,52)[8,u2][sd-C] | 37 | i-LBP(1,30)[8,u2][sd-B] |
| 14 | i-LBP(1,38)[8,u2][sd-A] | 38 | i-LBP(1,51)[8,u2][Max-A] |
| 15 | i-LBP(1,46)[8,u2][sd-C] | 39 | i-LBP(2,57)[8,u2][Max-B] |
| 16 | i-LBP(3,12)[8,u2][sd-C] | 40 | Fourier Ang (2,2)[rad][Max-C] |
| 17 | i-LBP(3,2)[8,u2][Max-A] | 41 | i-LBP(3,15)[8,u2][Max-A] |
| 18 | i-LBP(2,30)[8,u2][sd-A] | 42 | i-LBP(1,37)[8,u2][sd-A] |
| 19 | i-LBP(2,46)[8,u2][sd-A] | 43 | i-LBP(1,50)[8,u2][sd-B] |
| 20 | i-LBP(4,13)[8,u2][sd-B] | 44 | i-LBP(4,21)[8,u2][sd-C] |
| 21 | i-LBP(2,20)[8,u2][sd-B] | 45 | i-LBP(2,35)[8,u2][sd-A] |
| 22 | i-LBP(2,20)[8,u2][sd-A] | 46 | i-LBP(1,8)[8,u2][Max-C] |
| 23 | i-LBP(2,48)[8,u2][sd-C] | 47 | i-LBP(3,34)[8,u2][sd-B] |
| 24 | i-LBP(1,29)[8,u2][sd-B] | 48 | i-LBP(3,7)[8,u2][sd-B] |
| Network Architecture | Classification Accuracy | |
|---|---|---|
| Validation Accuracy (%) | Test Accuracy (%) | |
| Simple ConvNets | 88.7 | 84.5 |
| AlexNet | 92.1 | 86.4 |
| VGG-19 | 91.2 | 86.9 |
| ResNet-50 | 92.5 | 87.3 |
| ResNet-101 | 91.9 | 86.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, M.R.; Yasmin, J.; Park, E.; Kim, G.; Kim, M.S.; Wakholi, C.; Mo, C.; Cho, B.-K. Classification of Watermelon Seeds Using Morphological Patterns of X-ray Imaging: A Comparison of Conventional Machine Learning and Deep Learning. Sensors 2020, 20, 6753. https://doi.org/10.3390/s20236753
Ahmed MR, Yasmin J, Park E, Kim G, Kim MS, Wakholi C, Mo C, Cho B-K. Classification of Watermelon Seeds Using Morphological Patterns of X-ray Imaging: A Comparison of Conventional Machine Learning and Deep Learning. Sensors. 2020; 20(23):6753. https://doi.org/10.3390/s20236753
Chicago/Turabian StyleAhmed, Mohammed Raju, Jannat Yasmin, Eunsung Park, Geonwoo Kim, Moon S. Kim, Collins Wakholi, Changyeun Mo, and Byoung-Kwan Cho. 2020. "Classification of Watermelon Seeds Using Morphological Patterns of X-ray Imaging: A Comparison of Conventional Machine Learning and Deep Learning" Sensors 20, no. 23: 6753. https://doi.org/10.3390/s20236753
APA StyleAhmed, M. R., Yasmin, J., Park, E., Kim, G., Kim, M. S., Wakholi, C., Mo, C., & Cho, B.-K. (2020). Classification of Watermelon Seeds Using Morphological Patterns of X-ray Imaging: A Comparison of Conventional Machine Learning and Deep Learning. Sensors, 20(23), 6753. https://doi.org/10.3390/s20236753






