FRET-Based Aptasensor for the Selective and Sensitive Detection of Lysozyme
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation of Aptasensor
2.3. Preparation of Functionalized Flow Cell
2.4. Aptasensor Immobilization and Single-Molecule Imaging
2.5. Single-Molecule Data Analysis
3. Results and Discussion
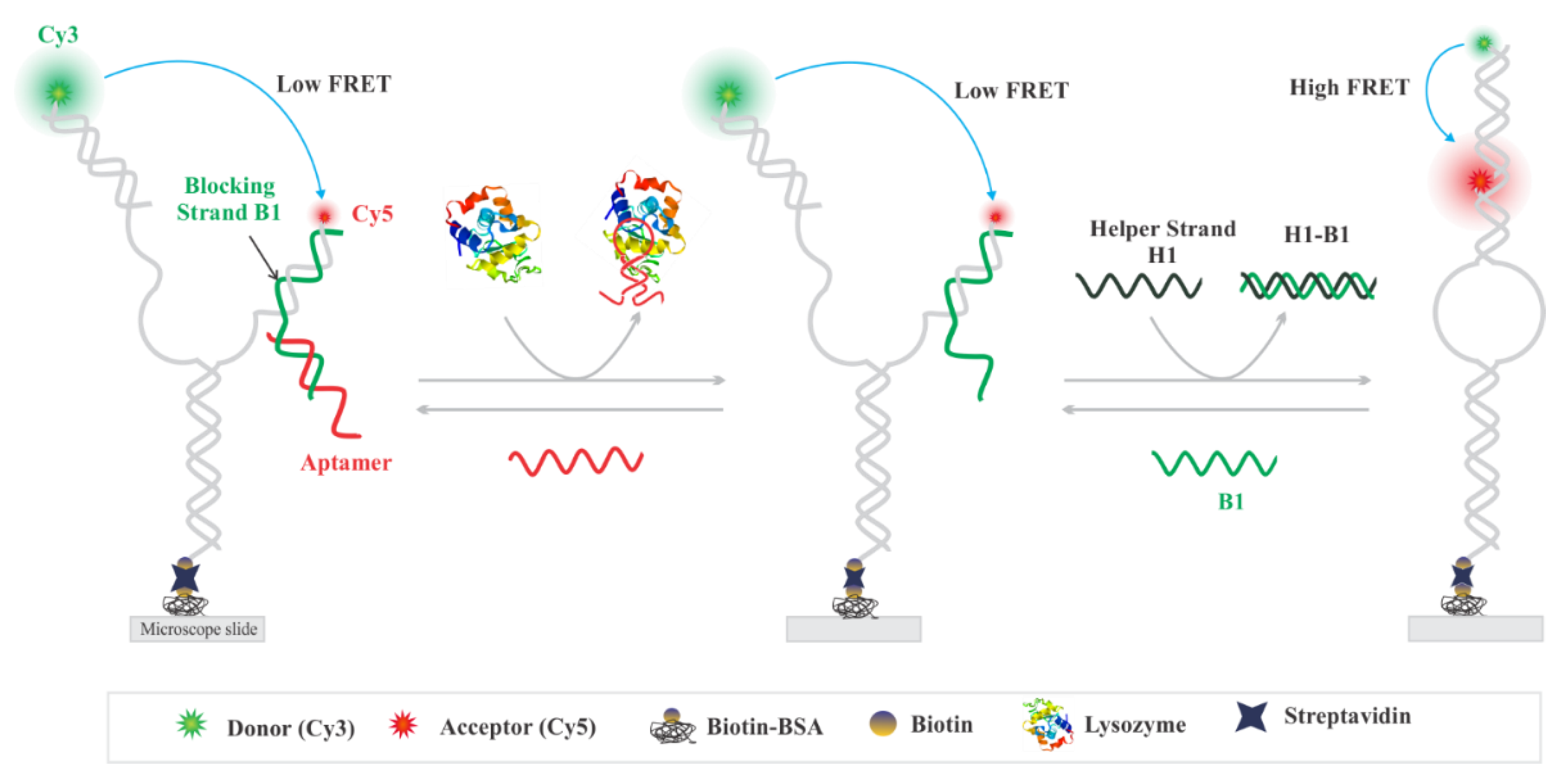
3.1. Experimental Design
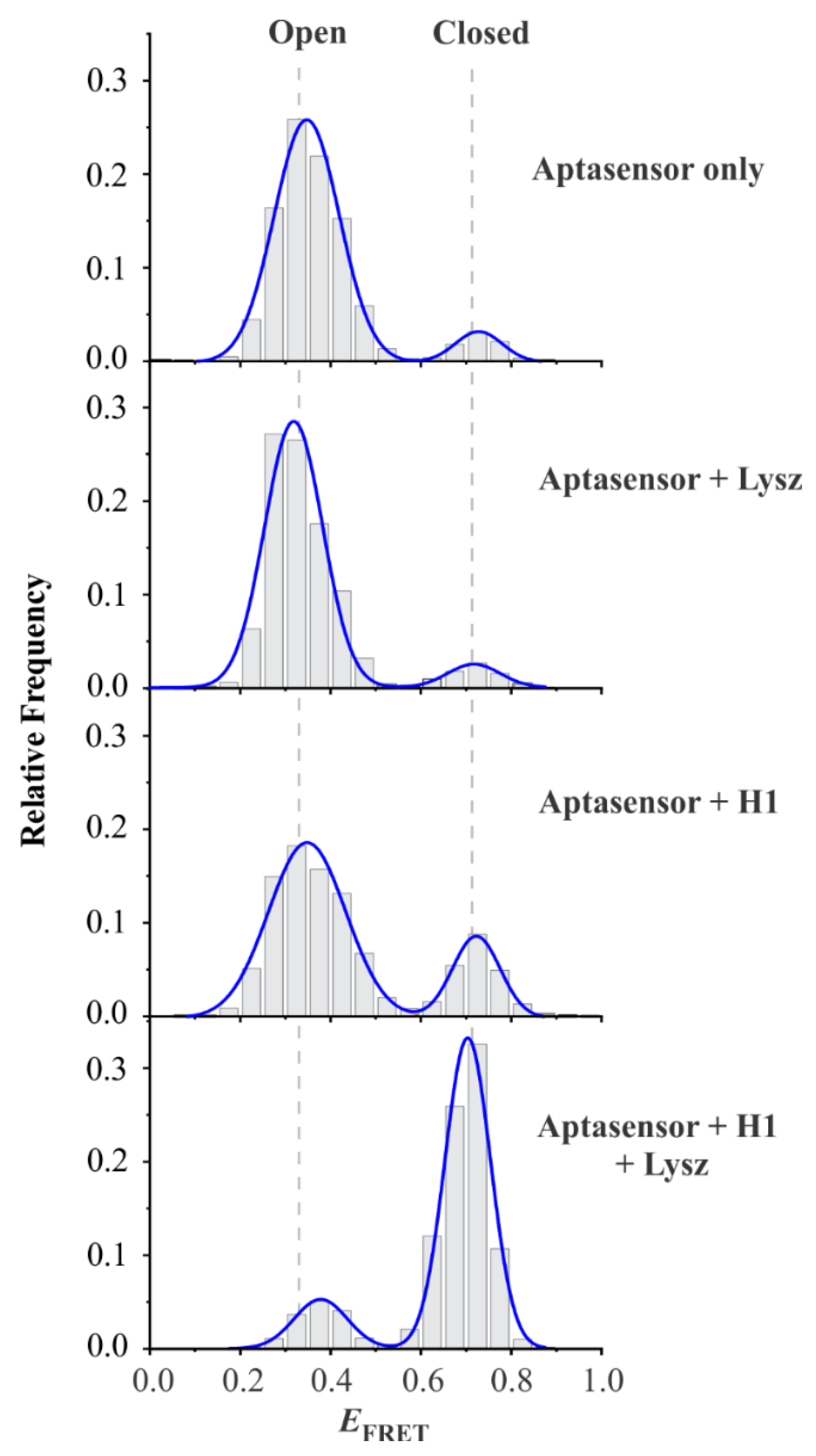
3.2. Single-Molecule Analysis of Aptasensor
3.3. Design Feasibility Assessment
3.4. Analytical Sensitivity
3.5. Selectivity of Lysozyme Aptasensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vasilescu, A.; Wang, Q.; Li, M.; Boukherroub, R.; Szunerits, S. Aptamer-Based Electrochemical Sensing of Lysozyme. Chemosensors 2016, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Jamei, H.R.; Rezaei, B.; Ensafi, A.A. An ultrasensitive electrochemical anti-lysozyme aptasensor with biorecognition surface based on aptamer/amino-rGO/ionic liquid/amino-mesosilica nanoparticles. Colloids Surf. B Biointerfaces 2019, 181, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Ragland, S.A.; Criss, A.K. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017, 13, e1006512. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.I.; Maddaus, A.G.; Song, E. A Low-Cost Inkjet-Printed Aptamer-Based Electrochemical Biosensor for the Selective Detection of Lysozyme. Biosensors 2018, 8, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Xia, N.; Li, T.; Bai, Y.; Chen, X. Aptasensor for visual and fluorometric determination of lysozyme based on the inner filter effect of gold nanoparticles on CdTe quantum dots. Microchim. Acta 2016, 183, 2917–2923. [Google Scholar] [CrossRef]
- Luo, R.; Zhou, X.; Chen, Y.; Tuo, S.; Jiang, F.; Niu, X.; Pan, F.; Wang, H. Lysozyme Aptamer-Functionalized Magnetic Nanoparticles for the Purification of Lysozyme from Chicken Egg White. Foods 2019, 8, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pushkaran, A.C.; Nataraj, N.; Nair, N.; Götz, F.; Biswas, R.; Mohan, C.G. Understanding the Structure–Function Relationship of Lysozyme Resistance in Staphylococcus aureusby Peptidoglycan O-Acetylation Using Molecular Docking, Dynamics, and Lysis Assay. J. Chem. Inf. Model. 2015, 55, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Dowarha, D.; Katte, R.; Chou, R.-H.; Filipek, A.; Yu, C. Lysozyme as the anti-proliferative agent to block the interaction between S100A6 and the RAGE V domain. PLoS ONE 2019, 14, e0216427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahanta, S.; Paul, S.; Srivastava, A.; Pastor, A.; Kundu, B.; Chaudhuri, T.K. Stable self-assembled nanostructured hen egg white lysozyme exhibits strong anti-proliferative activity against breast cancer cells. Colloids Surf. B Biointerfaces 2015, 130, 237–245. [Google Scholar] [CrossRef]
- Johansson, B.G.; Malmquist, J. Quantitative Immunochemical Determination of Lysozyme (Muramidase) in Serum and Urine. Scand. J. Clin. Lab. Investig. 1971, 27, 255–261. [Google Scholar] [CrossRef]
- Tomita, H.; Sato, S.; Matsuda, R.; Sugiura, Y.; Kawaguchi, H.; Niimi, T.; Yoshida, S.; Morishita, M. Serum lysozyme levels and clinical features of sarcoidosis. Lung 1999, 177, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Grieco, M.H.; Reddy, M.M.; Kothari, H.B.; Lange, M.; Buimovici-Klein, E.; William, D. Elevated β2-microglobulin and lysozyme levels in patients with acquired immune deficiency syndrome. Clin. Immunol. Immunopathol. 1984, 32, 174–184. [Google Scholar] [CrossRef]
- Serra, C.; Vizoso, F.; Alonso, L.; Rodríguez, J.C.; González, L.O.; Fernández, M.; Lamelas, M.L.; Sánchez, L.M.; García-Muñiz, J.L.; Baltasar, A.; et al. Expression and prognostic significance of lysozyme in male breast cancer. Breast Cancer Res. 2002, 4, R16. [Google Scholar] [CrossRef] [Green Version]
- Polimeni, M.; Valente, E.; Aldieri, E.; Khadjavi, A.; Giribaldi, G.; Prato, M. Human lysozyme as a potential diagnostic marker in malaria: A mechanistic study of haemozoin-induced monocyte degranulation. Malar. J. 2012, 11, P80. [Google Scholar] [CrossRef] [Green Version]
- Helmfors, L.; Boman, A.; Civitelli, L.; Nath, S.; Sandin, L.; Janefjord, C.; McCann, H.; Zetterberg, H.; Blennow, K.; Halliday, G.; et al. Protective properties of lysozyme on β-amyloid pathology: Implications for Alzheimer disease. Neurobiol. Dis. 2015, 83, 122–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandin, L.; Nath, S.; Armstrong, A.; Janefjord, C.; McCann, H.; Halliday, G.M.; Blennow, K.; Zetterberg, H.; Brorsson, A.-C.; Kagedal, K. The Role of Lysozyme in Alzheimer’s Disease. Alzheimers Dement 2015, 11, P477–P478. [Google Scholar] [CrossRef]
- Mishra, O.P.; Batra, P.; Ali, Z.; Anupurba, S.; Das, B.K. Cerebrospinal fluid lysozyme level for the diagnosis of tuberculous meningitis in children. J. Trop. Pediatr. 2003, 49, 13–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruzanski, W.; Saito, S.; Ogryzlo, M.A. The significance of lysozyme (muramidase) in rheumatoid arthritis. i. levels in serum and synovial fluid. Arthritis Rheum. 1970, 13, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Torsteinsdóttir, I.; Håkansson, L.; Hällgren, R.; Gudbjörnsson, B.; Arvidson, N.-G.; Venge, P. Serum lysozyme: A potential marker of monocyte/macrophage activity in rheumatoid arthritis. Rheumatology 1999, 38, 1249–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falchuk, K.R.; Perrotto, J.L.; Isselbacher, K.J. Serum Lysozyme in Crohn’s Disease. A Useful Index of Disease Activity. Gastroenterology 1975, 69, 893–896. [Google Scholar] [CrossRef]
- Daeschel, M.A.; Musafija-Jeknic, T.; Wu, Y.; Bizzarri, D.; Villa, A. High-Performance Liquid Chromatography Analysis of Lysozyme in Wine. Am. J. Enol. Vitic. 2002, 53, 154–157. [Google Scholar]
- Carstens, C.; Deckwart, M.; Webber-Witt, M.; Schäfer, V.; Eichhorn, L.; Brockow, K.; Fischer, M.; Christmann, M.; Paschke-Kratzin, A. Evaluation of the Efficiency of Enological Procedures on Lysozyme Depletion in Wine by an Indirect ELISA Method. J. Agric. Food Chem. 2014, 62, 6247–6253. [Google Scholar] [CrossRef] [PubMed]
- Kerkaert, B.; Mestdagh, F.; De Meulenaer, B. Detection of hen’s egg white lysozyme in food: Comparison between a sensitive HPLC and a commercial ELISA method. Food Chem. 2010, 120, 580–584. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, Q.; Tang, G.; Liu, S.; Xu, S.; Zhang, X. A facile electrochemical aptasensor for lysozyme detection based on target-induced turn-off of photosensitization. Biosens. Bioelectron. 2019, 126, 412–417. [Google Scholar] [CrossRef]
- Ortiz-Aguayo, D.; Del Valle, M. Label-Free Aptasensor for Lysozyme Detection Using Electrochemical Impedance Spectroscopy. Sensors 2018, 18, 354. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Gan, S.; Xu, Q.; Qiu, X.; Gao, P.; Huang, S. A three-way junction aptasensor for lysozyme detection. Biosens. Bioelectron. 2013, 39, 250–254. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.; Dong, Y.; Chu, X. Electrogenerated chemiluminescence aptasensor for lysozyme based on copolymer nanospheres encapsulated black phosphorus quantum dots. Talanta 2019, 199, 507–512. [Google Scholar] [CrossRef]
- Fang, M.; Zhuo, K.; Chen, Y.; Zhao, Y.; Bai, G.; Wang, J. Fluorescent probe based on carbon dots/silica/molecularly imprinted polymer for lysozyme detection and cell imaging. Anal. Bioanal. Chem. 2019, 411, 5799–5807. [Google Scholar] [CrossRef]
- Zuo, L.; Qin, G.; Lan, Y.; Wei, Y.; Dong, C. A turn-on phosphorescence aptasensor for ultrasensitive detection of lysozyme in humoral samples. Sens. Actuators B Chem. 2019, 289, 100–105. [Google Scholar] [CrossRef]
- Lou, T.; Qiang, H.; Chen, Z. Core-shell Cu@Au nanoparticles-based colorimetric aptasensor for the determination of lysozyme. Talanta 2017, 163, 132–139. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Yu, C.-J.; Cheng, T.-L.; Tseng, W.-L. Colorimetric Detection of Lysozyme Based on Electrostatic Interaction with Human Serum Albumin-Modified Gold Nanoparticles. Langmuir 2008, 24, 3654–3660. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, A.; Gáspár, S.; Gheorghiu, M.; David, S.; Dinca, V.; Peteu, S.; Wang, Q.; Li, M.; Boukherroub, R.; Szunerits, S. Surface Plasmon Resonance based sensing of lysozyme in serum on Micrococcus lysodeikticus-modified graphene oxide surfaces. Biosens. Bioelectron. 2017, 89, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Mihai, I.; Vezeanu, A.; Polonschii, C.; Albu, C.; Radu, G.-L.; Vasilescu, A. Label-free detection of lysozyme in wines using an aptamer based biosensor and SPR detection. Sens. Actuators B Chem. 2015, 206, 198–204. [Google Scholar] [CrossRef]
- Cheng, A.K.H.; Ge, B.; Yu, H.-Z. Aptamer-Based Biosensors for Label-Free Voltammetric Detection of Lysozyme. Anal. Chem. 2007, 79, 5158–5164. [Google Scholar] [CrossRef]
- Sekar, R.B.; Periasamy, A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J. Cell Boil. 2003, 160, 629–633. [Google Scholar] [CrossRef] [Green Version]
- Nimse, S.B.; Sonawane, M.D.; Song, K.-S.; Kim, T. Biomarker detection technologies and future directions. Analyst 2016, 141, 740–755. [Google Scholar] [CrossRef] [Green Version]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef]
- Chikkaveeraiah, B.V.; Bhirde, A.A.; Morgan, N.Y.; Eden, H.S.; Chen, X. Electrochemical Immunosensors for Detection of Cancer Protein Biomarkers. ACS Nano 2012, 6, 6546–6561. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Zu, Y. A Highlight of Recent Advances in Aptamer Technology and Its Application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Liu, J.; Chen, K.; Xu, Y.; Liu, B.; Liao, J.; Zhu, L.; Hu, X.; Li, J.; Pu, Y.; et al. Selection and characterization of DNA aptamer against glucagon receptor by cell-SELEX. Sci. Rep. 2017, 7, 7179. [Google Scholar] [CrossRef] [Green Version]
- Pei, X.; Zhang, J.; Liu, J. Clinical applications of nucleic acid aptamers in cancer. Mol. Clin. Oncol. 2014, 2, 341–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, T. Single-Molecule Fluorescence Resonance Energy Transfer. Methods 2001, 25, 78–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Yang, Y.; Zhang, C.-Y. Single-molecule FRET for Ultrasensitive Detection of Biomolecules. NanoBioImaging 2014, 1. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, D.R.; Kaur, A.; Megalathan, A.; Sapkota, K.; Dhakal, S. Build Your Own Microscope: Step-By-Step Guide for Building a Prism-Based TIRF Microscope. Methods Protoc. 2018, 1, 40. [Google Scholar] [CrossRef] [Green Version]
- Srinivas, N.; Ouldridge, T.E.; Šulc, P.; Schaeffer, J.M.; Yurke, B.; Louis, A.A.; Doye, J.P.K.; Winfree, E. On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic Acids Res. 2013, 41, 10641–10658. [Google Scholar] [CrossRef]
- Cox, J.; Ellington, A.D. Automated selection of anti-protein aptamers. Bioorganic Med. Chem. 2001, 9, 2525–2531. [Google Scholar] [CrossRef]
- Kaur, A.; Sapkota, K.; Dhakal, S. Multiplexed Nucleic Acid Sensing with Single-Molecule FRET. ACS Sens. 2019, 4, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Megalathan, A.; Cox, B.D.; Wilkerson, P.D.; Kaur, A.; Sapkota, K.; Reiner, J.E.; Dhakal, S. Single-molecule analysis of i-motif within self-assembled DNA duplexes and nanocircles. Nucleic Acids Res. 2019, 47, 7199–7212. [Google Scholar] [CrossRef] [Green Version]
- Sapkota, K.; Megalathan, A.; Moore, D.; Dhakal, S.; Kaur, A.; Donkoh-Moore, C. Single-Step FRET-Based Detection of Femtomoles DNA. Sensors 2019, 19, 3495. [Google Scholar] [CrossRef] [Green Version]
- Center for the Physics of Living Cells. smFRET Data Acquisition and Analysis Package. Available online: https://cplc.illinois.edu/software/ (accessed on 8 February 2020).
- Roy, R.; Hohng, S.; Ha, T. A Practical Guide to Single Molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef]
- Aitken, C.E.; Marshall, R.A.; Puglisi, J.D. An Oxygen Scavenging System for Improvement of Dye Stability in Single-Molecule Fluorescence Experiments. Biophys. J. 2008, 94, 1826–1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porstmann, B.; Jung, K.; Schmechta, H.; Evers, U.; Pergande, M.; Porstmann, T.; Kramm, H.-J.; Krause, H. Measurement of lysozyme in human body fluids: Comparison of various enzyme immunoassay techniques and their diagnostic application. Clin. Biochem. 1989, 22, 349–355. [Google Scholar] [CrossRef]
- Rubio-Ruiz, M.E.; Díaz-Díaz, E.; Cárdenas-León, M.; Argüelles-Medina, R.; Sánchez-Canales, P.; Larrea-Gallo, F.; Soria-Castro, E.; Guarner-Lans, V. Glycation does not modify bovine serum albumin (BSA)-induced reduction of rat aortic relaxation: The response to glycated and nonglycated BSA is lost in metabolic syndrome. Glycobiology 2008, 18, 517–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukumori, Y.; Takeda, H.; Fujisawa, T.; Ushijima, K.; Onodera, S.; Shiomi, N. Blood glucose and insulin concentrations are reduced in humans administered sucrose with inosine or adenosine. J. Nutr. 2000, 130, 1946–1949. [Google Scholar] [CrossRef] [Green Version]
- Eleftheriadis, T.; Pissas, G.; Liakopoulos, V.; Stefanidis, I. Cytochrome c as a Potentially Clinical Useful Marker of Mitochondrial and Cellular Damage. Front. Immunol. 2016, 7, 11. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapkota, K.; Dhakal, S. FRET-Based Aptasensor for the Selective and Sensitive Detection of Lysozyme. Sensors 2020, 20, 914. https://doi.org/10.3390/s20030914
Sapkota K, Dhakal S. FRET-Based Aptasensor for the Selective and Sensitive Detection of Lysozyme. Sensors. 2020; 20(3):914. https://doi.org/10.3390/s20030914
Chicago/Turabian StyleSapkota, Kumar, and Soma Dhakal. 2020. "FRET-Based Aptasensor for the Selective and Sensitive Detection of Lysozyme" Sensors 20, no. 3: 914. https://doi.org/10.3390/s20030914





