Diabetic Retinopathy Prediction by Ensemble Learning Based on Biochemical and Physical Data
Abstract
:1. Introduction
- A new wrapped feature selection algorithm, XGBIBS (Improved Backward Search Based on XGBoost), was proposed to reduce feature redundancy and improve the effect of a single ensemble learning classifier. The buffer feature subset was added to make it possible to operate on multiple features, and XGBIBS searches for the optimal subset in the sorting space based on the different feature metrics of XGBoost;
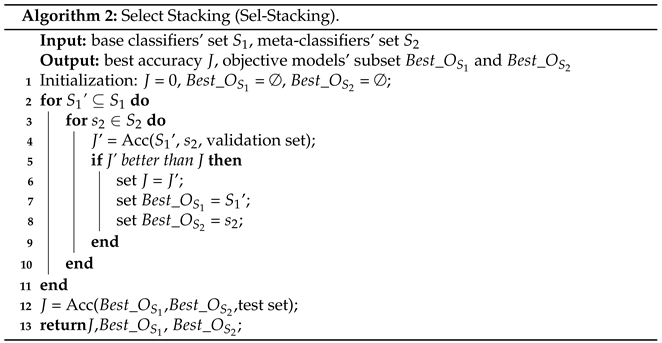
- A stacking model fusion method, Sel-Stacking (Select-Stacking), was proposed to improve the performance of a single model. There were two improvements to the algorithm. Sel-Stacking not only kept Label-Proba as the input matrix of the meta-classifier, but also determined the optimal combination of base classifiers by a global search;
- A diabetic retinopathy prediction model, XGB-Stacking, was constructed to predict the risk of diabetic retinopathy by combining the XGBIBS feature selection algorithm and the Sel-Stacking model fusion method.
2. Related Work
2.1. Prediction of Diabetic Retinopathy
2.2. Feature Selection Algorithm
2.2.1. Wrapped Feature Selection Based on Heuristic Search
2.2.2. Feature Selection Algorithm Based on XGBoost
2.3. Multi-Model by Stacking Ensemble Learning
3. Materials and Methods
3.1. Materials
3.2. XGBIBS Feature Selection
3.2.1. XGBoost Feature Importance Metrics in the XGBIBS Algorithm
- It filters out the features with zero importance. In tree segmentation, it is inevitable that features with a zero importance metric will appear. Most of the features with zero importance are not distinguishable from the samples, as their information value is very low. The XGBIBS feature selection algorithm can filter out the features with zero importance so that they cannot enter the feature search space;
- It outputs multiple different feature importance metrics at the same time. It is necessary to construct two ranking spaces for the sequence search according to certain rules. After XGBoost calculates the feature importance metrics, it outputs a variety of rankings to be used in the XGBIBS feature selection algorithm. Different feature importance metrics of the BFGSBS strategy can be chosen arbitrarily.
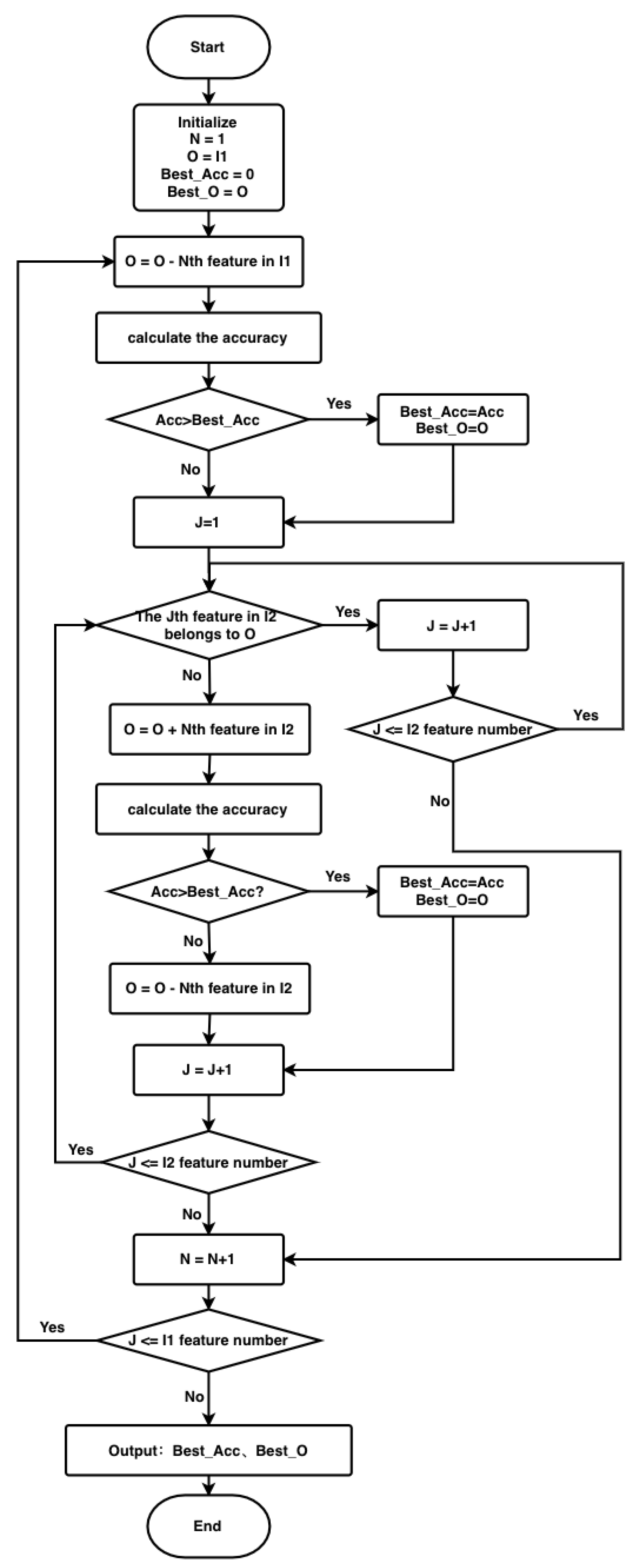
3.2.2. The BFGSBS Strategy in the XGBIBS Algorithm
- Delete the Nth feature from the buffer feature subset O each time (the starting value of N is 1) according to the feature importance queue , and the buffer feature subset O is updated;
- Use the new buffer feature subset O to calculate the evaluation function. If the result is better than that of the optimal evaluation function, save this buffer feature subset as a new optimal feature subset ;
- After this round of operation, , and go to Stage 2Stage 2 is the floating forward increase:
- Search for a feature that is not in the buffer feature subset O and in turn from the feature importance queue ;
- If this feature is added to the buffer feature subset O, the effect of the evaluation function is improved. Then, the buffer feature subset O is updated, and the buffer feature subset is saved as a new optimal feature subset, ;
- End this stage after traversing the order from beginning to end, and return to Stage 1.
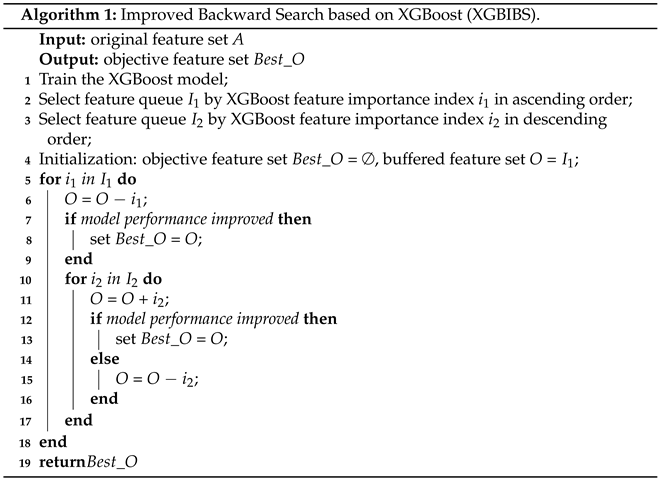
3.2.3. Time Complexity of the XGBIBS Algorithm
3.3. Multi-Model by Sel-Stacking Ensemble Learning
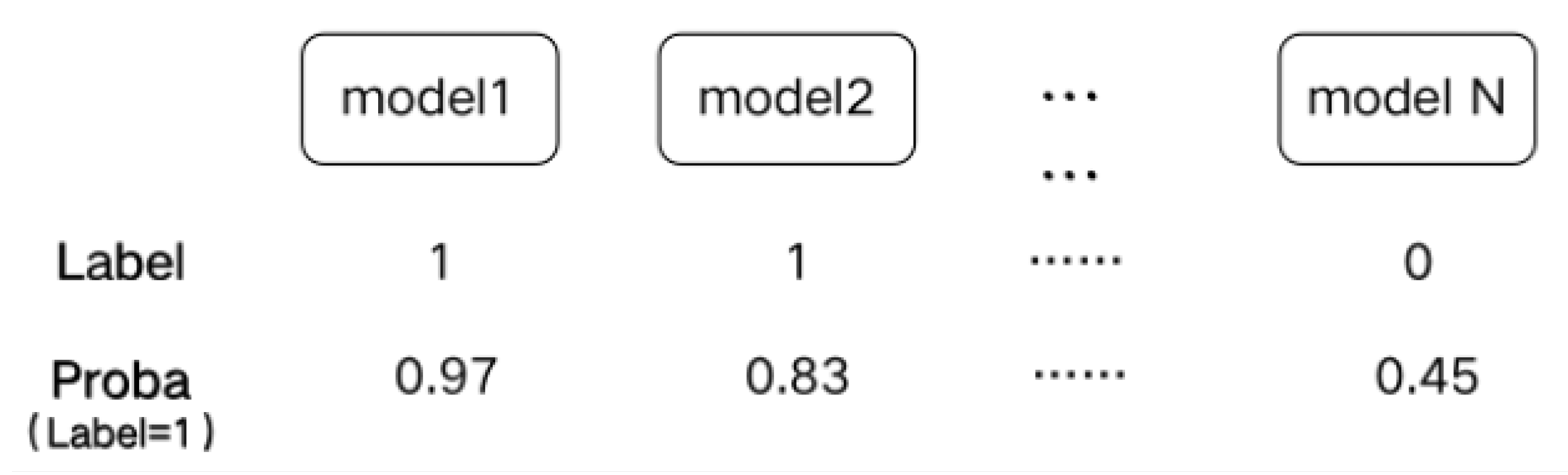
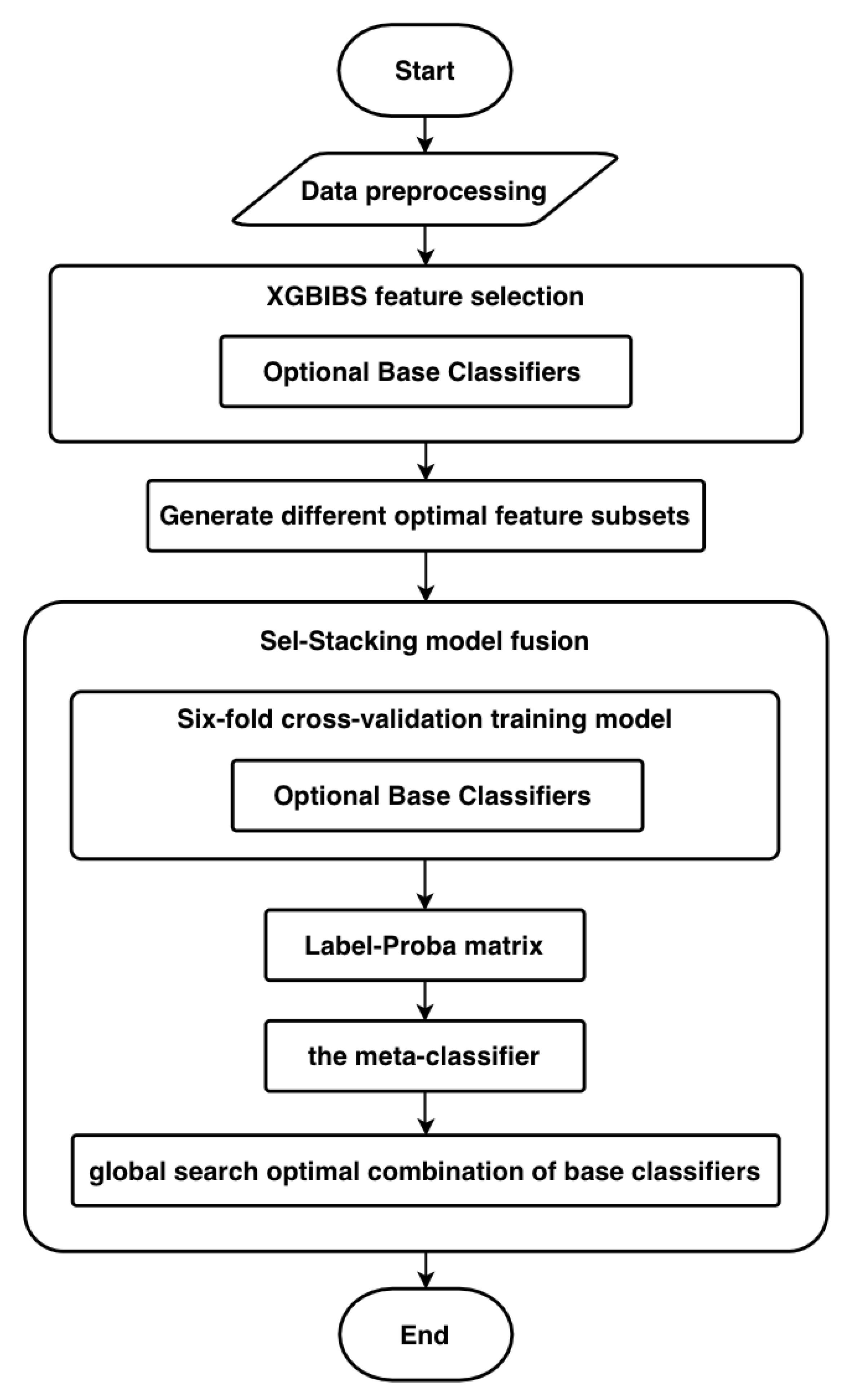
- The Sel-Stacking method changes the input of meta-classifiers. In order to avoid overfitting, the output label and Proba of the base classifiers are retained and used as the input of the meta-classifier at the same time. This prediction is a binary classification problem. A single learner outputs the predicted label and the corresponding classification probability Proba. The Label-Proba matrix predicted by one base classifier of one input sample is shown in Figure 3. For each sample, N base classifiers produce a 2*N output matrix, and M samples with N base classifiers produce a 2M*N Label-Proba matrix.
- The Sel-Stacking method improves the combination of learners and selects the optimal combination based on the data. For the model of the base classifiers, a variety of classifiers that have applied the XGBIBS feature selection algorithm were respectively connected, and the model was trained with six-fold cross-validation. The Sel-Stacking method adds a feature selection process between the base classifiers and the meta-classifier through a global search to select the best set of base classifiers.
3.4. XGB-Stacking Model Based on the XGBIBS Algorithm and the Sel-Stacking Method
3.5. Performance Evaluation Matrix
4. Experiments and Results Discussion
4.1. Experimental Setup
4.1.1. Experimental Environment
4.1.2. Dataset Partition
4.1.3. Classifiers’ Selection
4.2. Experimental Results of the XGBIBS Feature Selection Algorithm
4.2.1. The Experimental Result of Using XGBoost Feature Importance Metrics
4.2.2. The Experimental Result of the BFGSBS Search Strategy
4.2.3. The Experimental Results of Different Feature Selection Algorithms
4.3. The Experimental Results of Model Fusion by Sel-Stacking
4.3.1. The Experimental Results of the Meta-Classifier’s Different Input Strategies
4.3.2. The Experimental Results of Different Classifier Combination Strategies
- Stacking A: KNN, AdaBoost, GBDT, XGBoost, LightGBM, and CatBoost (all optional classifiers chosen as base classifiers in the stacking method);
- Stacking B: AdaBoost, GBDT, XGBoost, LightGBM, and CatBoost (base classifiers Combination A by selecting randomly in the stacking method);
- Stacking C: GBDT, XGBoost, LightGBM, and CatBoost (base classifiers Combination B by selecting randomly in the stacking method);
- Voting A: KNN, AdaBoost, GBDT, XGBoost, LightGBM, and CatBoost (voting method using all optional classifiers);
- Voting B: KNN, GBDT, XGBoost, and CatBoost (voting method using the base classifiers chosen by the Sel-Stacking method);
- Blending A: KNN, AdaBoost, GBDT, XGBoost, LightGBM, and CatBoost (blending method using all optional classifiers);
- Blending B: KNN, GBDT, XGBoost, and CatBoost (blending method using the base classifiers chosen by the Sel-Stacking method).
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| XGBIBS | Improved Backward Search Based on XGBoost |
| BFGSBS | Buffer Floating Generalized Sequential Backward Search |
| SFBS | Sequential Floating Backward Search |
| ISFFS | Improved Sequential Floating Forward Selection |
References
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karurangaa, S.; Unwinb, N.; Colagiuric, S.; Guariguatad, L.; Motalae, A.; Ogurtsovaf, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abràmoff, M.D.; Reinhardt, J.M.; Russell, S.R.; Folk, J.C.; Mahajan, V.B.; Niemeijer, M.; Quellec, G. Automated early detection of diabetic retinopathy. Ophthalmology 2010, 117, 1147–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahlsten, J.; Jaskari, J.; Kivinen, J.; Turunen, L.; Jaanio, E.; Hietala, K.; Kaski, K. Deep learning fundus image analysis for diabetic retinopathy and macular edema grading. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadekallu, T.R.; Khare, N.; Bhattacharya, S.; Singh, S.; Maddikunta, P.K.R.; Srivastava, G. Deep neural networks to predict diabetic retinopathy. J. Ambient. Intell. Humaniz. Comput. 2020, 1–14. [Google Scholar] [CrossRef]
- Somasundaram, S.K.; Alli, P. A machine learning ensemble classifier for early prediction of diabetic retinopathy. J. Med. Syst. 2017, 41, 1–12. [Google Scholar]
- Tsao, H.Y.; Chan, P.Y.; Su, E.C.Y. Predicting diabetic retinopathy and identifying interpretable biomedical features using machine learning algorithms. BMC Bioinform. 2018, 19, 111–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietterich, T.G. Ensemble Methods in Machine Learning. In International Workshop on Multiple Classifier Systems; Springer: Berlin/Heidelberg, Germany, 2000. [Google Scholar]
- Seni, G.; Elder, J.F. Ensemble methods in data mining: Improving accuracy through combining predictions. Synth. Lect. Data Min. Knowl. Discov. 2010, 2, 1–126. [Google Scholar] [CrossRef]
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3146–3154. [Google Scholar]
- Dorogush, A.V.; Ershov, V.; Gulin, A. CatBoost: Gradient boosting with categorical features support. arXiv 2018, arXiv:1810.11363. [Google Scholar]
- Ramani, R.G.; Balasubramanian, L.; Jacob, S.G. Automatic prediction of Diabetic Retinopathy and Glaucoma through retinal image analysis and data mining techniques. In Proceedings of the International Conference on Machine Vision and Image Processing (MVIP), Coimbatore, India, 14–15 December 2012; pp. 149–152. [Google Scholar]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Z. Feature selection algorithm based on XGBoost. J. Commun. 2019, 40, 1–8. [Google Scholar]
- Tan, F.; Fu, X.; Zhang, Y.; Bourgeois, A.G. A genetic algorithm-based method for feature subset selection. Soft Comput. 2008, 12, 111–120. [Google Scholar] [CrossRef]
- Nakariyakul, S.; Casasent, D.P. An improvement on floating search algorithms for feature subset selection. Pattern Recognit. 2009, 42, 1932–1940. [Google Scholar] [CrossRef]
- Fallahpour, S.; Lakvan, E.N.; Zadeh, M.H. Using an ensemble classifier based on sequential floating forward selection for financial distress prediction problem. J. Retail. Consum. Serv. 2017, 34, 159–167. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Sang, X.; Xiao, W.; Zheng, H.; Yang, Y.; Liu, T. HMMPred: Accurate Prediction of DNA-binding proteins based on HMM Profiles and XGBoost feature selection. Comput. Math. Methods Med. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Zhang, Q.; Yu, B.; Yu, Z.; Lawrence, P.J.; Ma, Q.; Zhang, Y. Improving protein-protein interactions prediction accuracy using XGBoost feature selection and stacked ensemble classifier. Comput. Biol. Med. 2020, 123, 103899. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, D.; Geng, N.; Wang, Y.; Yin, Y.; Jin, Y. Stacking-based ensemble learning of decision trees for interpretable prostate cancer detection. Appl. Soft Comput. 2019, 77, 188–204. [Google Scholar] [CrossRef]
- Rawat, R.M.; Akram, M.; Pradeep, S.S. Dementia Detection Using Machine Learning by Stacking Models. In Proceedings of the 2020 5th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 10–12 June 2020; pp. 849–854. [Google Scholar]
- Hammam, A.A.; Elmousalami, H.H.; Hassanien, A.E. Stacking Deep Learning for Early COVID-19 Vision Diagnosis. In Big Data Analytics and Artificial Intelligence against COVID-19: Innovation Vision and Approach; Springer: Cham, Switzerland, 2020; pp. 297–307. [Google Scholar]
- Ji, S.; Li, R.; Shen, S.; Li, B.; Zhou, B.; Wang, Z. Heartbeat Classification Based on Multifeature Combination and Stacking-DWKNN Algorithm. J. Healthc. Eng. 2021. [Google Scholar] [CrossRef] [PubMed]
| Feature Category | Number | Features |
|---|---|---|
| Basic Information | 9 | NATION, MARITAL_STATUS, SEX, AGE, BMI, BP_HIGH, BP_LOW, HEIGHT, WEIGHT |
| Disease Information | 32 | A_S, ARRHYTHMIAS, CHD, MI, LUNG_TUMOR, BILIARY_TRACT_DISEASE, CHF, CIRRHOSIS, BREAST_TUMOR, CAROTID_ARTERY_STENOSIS, CEREBRAL_APOPLEXTY, CLD, DIGESTIVE_CARCINOMA, ENDOCRINE_DISEASE, FLD, GYNECOLGICAL_TUMOR, HEMATONOSIS, HYPERLIPIDEMIA, LEADDP, HYPERTENTION, INTRACRANIAL_TUMOR, OTHER_TUMOR, MEN, NEPHROPATHY, PCOS, NERVOUS_SYSTEM_DISEASE, PREGNANT, PANCREATIC_DISEASE, RENAL_FALIURE, RESPIRATORY_SYSTEM_DISEASE, RHEUMATIC_IMMUNITY, UROLOGIC_NEOPLASMS |
| Biochemical Indicators | 27 | ALB, ALP, ALT, AST, DBILI, GGT, GLO, IBILI, LDH_L, TBILI, TP, BU, SCR, SUA, HDL_C, LDL_C, TC, TG, HB, PCV, PLT, GLU, HBA1C, APTT, FBG, PT, PTA |
| SEX | Number of Samples | Percentage |
|---|---|---|
| Male (SEX = 1) | 1869 | 62.51% |
| Female (SEX = 0) | 1121 | 37.49% |
| AGE | Number of Samples | Percentage |
|---|---|---|
| Under 20 years old | 2 | 0.07% |
| 21–30 | 38 | 1.27% |
| 31–40 | 139 | 4.65% |
| 41–50 | 562 | 18.80% |
| 51–60 | 1018 | 34.05% |
| 61–70 | 860 | 28.76% |
| 71 years old and above | 371 | 12.41% |
| Features | Percentage (Value = 0) | Percentage (Value = 1) |
|---|---|---|
| NATION | 95.65% | 5.35% |
| MARITAL_STATUS | 97.89% | 2.11% |
| Parameter | Description |
|---|---|
| weight | the number of times a feature is used to split the data across all trees |
| gain | the average gain of the feature when it is used in trees |
| cover | the average coverage of the feature when it is used in trees |
| total_gain | the total gain of the feature when it is used in trees |
| total_cover | the total coverage of the feature when it is used in trees |
| Symbol | Definition |
|---|---|
| Queue arranged from small to large according to the importance of the XGBoost model | |
| Queue arranged from large to small according to the importance of the XGBoost model | |
| O | The buffer feature subset |
| The optimal feature subset | |
| Classification accuracy | |
| The highest classification accuracy | |
| N | Traverse the control variables of the queue |
| J | Traverse the control variables of the queue |
| Feature | Ranking | Score |
|---|---|---|
| NEPHROPATHY | 1 | 19.10 |
| HEIGHT | 2 | 4.95 |
| HBA1C | 3 | 3.69 |
| CHD | 4 | 3.57 |
| LEADDP | 5 | 3.40 |
| OTHER_TUMOR | 6 | 3.24 |
| RESPIRATORY_SYSTEM_DISEASE | 7 | 3.10 |
| RENAL_FALIURE | 8 | 2.69 |
| HYPERLIPIDEMIA | 9 | 2.48 |
| GYNECOLGICAL_TUMORHEIGHT | 10 | 2.25 |
| Classifier | NoFS (%) | BFGSBS1 (%) | BFGSBS2 (%) | BFGSBS (%) |
|---|---|---|---|---|
| KNN | 66.89 | 69.90 | 69.23 | 75.08 |
| AdaBoost | 78.09 | 80.60 | 80.43 | 80.60 |
| GBDT | 79.43 | 81.10 | 80.60 | 82.27 |
| XGBoost | 76.76 | 82.27 | 81.44 | 82.27 |
| LightGBM | 79.10 | 81.77 | 81.11 | 81.77 |
| CatBoost | 80.27 | 82.78 | 82.94 | 83.11 |
| Classifier | NoFS (%) | BFGSBS1 (%) | BFGSBS2 (%) | BFGSBS (%) |
|---|---|---|---|---|
| KNN | 0 | 69.12 | 79.41 | 82.35 |
| AdaBoost | 0 | 64.71 | 47.06 | 64.71 |
| GBDT | 0 | 20.59 | 54.41 | 42.65 |
| XGBoost | 0 | 42.65 | 32.35 | 42.65 |
| LightGBM | 0 | 44.12 | 47.06 | 38.24 |
| CatBoost | 0 | 51.47 | 41.18 | 51.47 |
| Classifier | NoFS (%) | SFBS (%) | ISFFS (%) | BFGSBS (%) |
|---|---|---|---|---|
| KNN | 66.89 | 71.07 | 77.93 | 75.08 |
| AdaBoost | 78.09 | 80.10 | 80.10 | 80.60 |
| GBDT | 79.43 | 81.61 | 76.92 | 82.27 |
| XGBoost | 76.76 | 80.77 | 77.93 | 82.27 |
| LightGBM | 79.10 | 81.10 | 75.08 | 81.77 |
| CatBoost | 80.27 | 82.44 | 77.42 | 83.11 |
| Classifier | NoFS (%) | SFBS (%) | ISFFS (%) | BFGSBS (%) |
|---|---|---|---|---|
| KNN | 0 | 25.00 | 88.24 | 82.35 |
| AdaBoost | 0 | 23.53 | 79.41 | 64.71 |
| GBDT | 0 | 22.06 | 85.29 | 42.65 |
| XGBoost | 0 | 20.59 | 85.29 | 42.65 |
| LightGBM | 0 | 19.12 | 85.29 | 38.24 |
| CatBoost | 0 | 82.35 | 82.35 | 51.47 |
| Classifier | GA (%) | SVM-RFE (%) | XGBIBS (%) |
|---|---|---|---|
| KNN | 73.24 | 72.07 | 75.08 |
| AdaBoost | 80.10 | 78.43 | 80.60 |
| GBDT | 81.77 | 80.94 | 82.77 |
| XGBoost | 81.27 | 81.94 | 82.77 |
| LightGBM | 82.27 | 81.10 | 81.77 |
| CatBoost | 81.77 | 83.11 | 83.11 |
| Classifier | GA (%) | SVM-RFE (%) | XGBIBS (%) |
|---|---|---|---|
| KNN | 55.88 | 33.82 | 82.35 |
| AdaBoost | 36.76 | 50.00 | 64.71 |
| GBDT | 36.76 | 57.35 | 42.65 |
| XGBoost | 44.12 | 48.53 | 42.65 |
| LightGBM | 51.47 | 55.88 | 38.24 |
| CatBoost | 47.06 | 22.06 | 51.47 |
| Feature Selection | Time (s) |
|---|---|
| NoFS | 0.17 |
| SVM-RFE | 40.05 |
| GA | 827.28 |
| BFGSBS | 65.20 |
| Input Strategy of the Meta-Classifier | Accuracy (%) |
|---|---|
| Label | 83.11 |
| Proba | 81.94 |
| Label-Proba | 83.95 |
| Method | Accuracy (%) | Time (s) |
|---|---|---|
| Sel-Stacking | 83.95 | 101.74 |
| Stacking A | 83.78 | 75.53 |
| Stacking B | 83.61 | 65.81 |
| Stacking C | 83.11 | 74.23 |
| Voting A | 83.44 | 8.79 |
| Voting B | 82.27 | 8.42 |
| Blending A | 80.10 | 8.49 |
| Blending B | 80.10 | 7.30 |
| Algorithm Category | Algorithm | Accuracy (%) |
|---|---|---|
| model fusion method | Sel-Stacking | 83.95 |
| Single classifier | KNN | 75.08 |
| AdaBoost | 80.60 | |
| GBDT | 82.77 | |
| XGBoost | 82.27 | |
| LightGBM | 81.77 | |
| CatBoost | 83.11 | |
| SVM | 74.25 | |
| LR | 74.91 | |
| Random Forest | 76.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Z.; Wu, Q.; Wang, Z.; Chen, G.; Lin, B. Diabetic Retinopathy Prediction by Ensemble Learning Based on Biochemical and Physical Data. Sensors 2021, 21, 3663. https://doi.org/10.3390/s21113663
Shen Z, Wu Q, Wang Z, Chen G, Lin B. Diabetic Retinopathy Prediction by Ensemble Learning Based on Biochemical and Physical Data. Sensors. 2021; 21(11):3663. https://doi.org/10.3390/s21113663
Chicago/Turabian StyleShen, Zun, Qingfeng Wu, Zhi Wang, Guoyi Chen, and Bin Lin. 2021. "Diabetic Retinopathy Prediction by Ensemble Learning Based on Biochemical and Physical Data" Sensors 21, no. 11: 3663. https://doi.org/10.3390/s21113663
APA StyleShen, Z., Wu, Q., Wang, Z., Chen, G., & Lin, B. (2021). Diabetic Retinopathy Prediction by Ensemble Learning Based on Biochemical and Physical Data. Sensors, 21(11), 3663. https://doi.org/10.3390/s21113663






