Biomimetic Sensors to Detect Bioanalytes in Real-Life Samples Using Molecularly Imprinted Polymers: A Review
Abstract
:1. Introduction
2. Medical Applications
2.1. Prevention and (Early) Diagnosis of Diseases
2.2. Pathogen Detection
2.3. Therapeutic Drug Monitoring
3. Food Safety
3.1. Mycotoxins
3.2. Pesticides
3.3. Bacteria Contamination
3.4. Food Adulteration
4. Environmental Monitoring
5. Drugs of Abuse and Their Precursors
6. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Polyakov, M. Adsorption properties and structure of silica gel. Zhur. Fiz. Khim. 1931, 2, 799–805. [Google Scholar]
- Belbruno, J.J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119, 94–119. [Google Scholar] [CrossRef] [PubMed]
- Refaat, D.; Aggour, M.G.; Farghali, A.A.; Mahajan, R.; Wiklander, J.G.; Nicholls, I.A.; Piletsky, S.A. Strategies for molecular imprinting and the evolution of MIP nanoparticles as plastic antibodies—synthesis and applications. Int. J. Mol. Sci. 2019, 20, 6304. [Google Scholar] [CrossRef] [Green Version]
- Panagiotopoulou, M.; Beyazit, S.; Nestora, S.; Haupt, K.; Tse Sum Bui, B. Initiator-free synthesis of molecularly imprinted polymers by polymerization of self-initiated monomers. Polymer 2015, 66, 43–51. [Google Scholar] [CrossRef]
- Erdőssy, J.; Horváth, V.; Yarman, A.; Scheller, F.W.; Gyurcsányi, R.E. Electrosynthesized molecularly imprinted polymers for protein recognition. TrAC Trends Anal. Chem. 2016, 79, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Crapnell, R.D.; Hudson, A.; Foster, C.W.; Eersels, K.; van Grinsven, B.; Cleij, T.J.; Banks, C.E.; Peeters, M. Recent advances in electrosynthesized molecularly imprinted polymer sensing platforms for bioanalyte detection. Sensors 2019, 19, 1204. [Google Scholar] [CrossRef] [Green Version]
- Beyazit, S.; Tse Sum Bui, B.; Haupt, K.; Gonzato, C. Molecularly imprinted polymer nanomaterials and nanocomposites by controlled/living radical polymerization. Prog. Polym. Sci. 2016, 62, 1–21. [Google Scholar] [CrossRef]
- Moein, M.M.; Abdel-Rehim, A.; Abdel-Rehim, M. Recent applications of molecularly imprinted sol-gel methodology in sample preparation. Molecules 2019, 24, 2889. [Google Scholar] [CrossRef] [Green Version]
- Branger, C.; Meouche, W.; Margaillan, A. Recent advances on ion-imprinted polymers. React. Funct. Polym. 2013, 73, 859–875. [Google Scholar] [CrossRef]
- Mujahid, A.; Aigner, S.; Dickert, F.L. Micro-structured interdigital capacitors with synthetic antibody receptors for ABO blood-group typing: Dedicated to Prof. Dr. Reinhard Niessner and Prof. Dr. Dietmar Knopp on the occasion of their 65th birthday. Sensors Actuators B Chem. 2017, 242, 378–383. [Google Scholar] [CrossRef]
- Turiel, E.; Martín-Esteban, A. Molecularly imprinted polymers-based microextraction techniques. TrAC Trends Anal. Chem. 2019, 118, 574–586. [Google Scholar] [CrossRef]
- He, C.; Long, Y.; Pan, J.; Li, K.; Liu, F. Application of molecularly imprinted polymers to solid-phase extraction of analytes from real samples. J. Biochem. Biophys. Methods 2007, 70, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Wulff, G. Enzyme-like catalysis by molecularly imprinted polymers. Chem. Rev. 2002, 102, 1–28. [Google Scholar] [CrossRef]
- Resmini, M. Molecularly imprinted polymers as biomimetic catalysts. Anal. Bioanal. Chem. 2012, 402, 3021–3026. [Google Scholar] [CrossRef]
- Muratsugu, S.; Shirai, S.; Tada, M. Recent progress in molecularly imprinted approach for catalysis. Tetrahedron Lett. 2020, 61, 151603. [Google Scholar] [CrossRef]
- Sanadgol, N.; Wackerlig, J. Developments of smart drug-delivery systems based on magnetic molecularly imprinted polymers for targeted cancer therapy: A short review. Pharmaceutics 2020, 12, 831. [Google Scholar] [CrossRef] [PubMed]
- Luliński, P. Molecularly imprinted polymers based drug delivery devices: A way to application in modern pharmacotherapy. A review. Mater. Sci. Eng. C 2017, 76, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Ramanavicius, S.; Jagminas, A.; Ramanavicius, A. Advances in molecularly imprinted polymers based affinity sensors (review). Polymers 2021, 13, 974. [Google Scholar] [CrossRef]
- Eersels, K.; Lieberzeit, P.; Wagner, P. A Review on Synthetic Receptors for Bioparticle Detection Created by Surface-Imprinting Techniques—From Principles to Applications. ACS Sensors 2016, 1, 1171–1187. [Google Scholar] [CrossRef]
- Ertürk, G.; Mattiasson, B. Molecular imprinting techniques used for the preparation of biosensors. Sensors 2017, 17, 288. [Google Scholar] [CrossRef] [Green Version]
- Unger, C.; Lieberzeit, P.A. Molecularly imprinted thin film surfaces in sensing: Chances and challenges. React. Funct. Polym. 2021, 161, 104855. [Google Scholar] [CrossRef]
- McKeating, K.S.; Aubé, A.; Masson, J.-F. Biosensors and nanobiosensors for therapeutic drug and response monitoring. Analyst 2016, 141, 429–449. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, D.; Weng, L.; Wang, L. Early lung cancer diagnosis by biosensors. Int. J. Mol. Sci. 2013, 14, 15479–15509. [Google Scholar] [CrossRef] [PubMed]
- Brazaca, L.C.; Sampaio, I.; Zucolotto, V.; Janegitz, B.C. Applications of biosensors in Alzheimer’s disease diagnosis. Talanta 2020, 210, 120644. [Google Scholar] [CrossRef]
- Kumar, M.S.; Ghosh, S.; Nayak, S.; Das, A.P. Recent advances in biosensor based diagnosis of urinary tract infection. Biosens. Bioelectron. 2016, 80, 497–510. [Google Scholar] [CrossRef]
- Templier, V.; Roupioz, Y. On the challenges of detecting whole Staphylococcus aureus cells with biosensors. J. Appl. Microbiol. 2017, 123, 1056–1067. [Google Scholar] [CrossRef] [Green Version]
- Altintas, Z. Biosensors and Nanotechnology: Applications in Health Care Diagnostics; John Wiley & Sons Press: Hoboken, NJ, USA, 2018; ISBN 9781119065036. [Google Scholar]
- Pirzada, M.; Sehit, E.; Altintas, Z. Cancer biomarker detection in human serum samples using nanoparticle decorated epitope-mediated hybrid MIP. Biosens. Bioelectron. 2020, 166, 112464. [Google Scholar] [CrossRef]
- Horikawa, R.; Sunayama, H.; Kitayama, Y.; Takano, E.; Takeuchi, T. A Programmable Signaling Molecular Recognition Nanocavity Prepared by Molecular Imprinting and Post-Imprinting Modifications. Angew. Chemie Int. Ed. 2016, 55, 13023–13027. [Google Scholar] [CrossRef]
- Mori, K.; Hirase, M.; Morishige, T.; Takano, E.; Sunayama, H.; Kitayama, Y.; Inubushi, S.; Sasaki, R.; Yashiro, M.; Takeuchi, T. A Pretreatment-Free, Polymer-Based Platform Prepared by Molecular Imprinting and Post-Imprinting Modifications for Sensing Intact Exosomes. Angew. Chemie Int. Ed. 2019, 58, 1612–1615. [Google Scholar] [CrossRef]
- Chunta, S.; Boonsriwong, W.; Wattanasin, P.; Naklua, W.; Lieberzeit, P.A. Direct assessment of very-low-density lipoprotein by mass sensitive sensor with molecularly imprinted polymers. Talanta 2021, 221, 121549. [Google Scholar] [CrossRef]
- Chunta, S.; Suedee, R.; Lieberzeit, P.A. Low-Density Lipoprotein Sensor Based on Molecularly Imprinted Polymer. Anal. Chem. 2016, 88, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Chunta, S.; Suedee, R.; Lieberzeit, P.A. High-density lipoprotein sensor based on molecularly imprinted polymer. Anal. Bioanal. Chem. 2018, 410, 875–883. [Google Scholar] [CrossRef]
- Chunta, S.; Suedee, R.; Boonsriwong, W.; Lieberzeit, P.A. Biomimetic sensors targeting oxidized-low-density lipoprotein with molecularly imprinted polymers. Anal. Chim. Acta 2020, 1116, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, Z.; Erdőssy, J.; Keltai, K.; Scheller, F.W.F.W.; Gyurcsányi, R.E.R.E. Electrosynthesized molecularly imprinted polyscopoletin nanofilms for human serum albumin detection. Anal. Chim. Acta 2017, 977, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Yu, Y.; Guo, M.; Lin, B.; Zhang, L. A sensitive determination of albumin in urine by molecularly imprinted electrochemical biosensor based on dual-signal strategy. Sensors Actuators B Chem. 2019, 288, 564–570. [Google Scholar] [CrossRef]
- Di Giulio, T.; Mazzotta, E.; Malitesta, C. Molecularly Imprinted Polyscopoletin for the Electrochemical Detection of the Chronic Disease Marker Lysozyme. Biosensors 2020, 11, 3. [Google Scholar] [CrossRef]
- Nontawong, N.; Amatatongchai, M.; Jarujamrus, P.; Nacapricha, D.; Lieberzeit, P.A. Novel dual-sensor for creatinine and 8-hydroxy-2′-deoxyguanosine using carbon-paste electrode modified with molecularly imprinted polymers and multiple-pulse amperometry. Sensors Actuators B Chem. 2021, 334, 129636. [Google Scholar] [CrossRef]
- Amatatongchai, M.; Sitanurak, J.; Sroysee, W.; Sodanat, S.; Chairam, S.; Jarujamrus, P.; Nacapricha, D.; Lieberzeit, P.A. Highly sensitive and selective electrochemical paper-based device using a graphite screen-printed electrode modified with molecularly imprinted polymers coated Fe3O4@Au@SiO2 for serotonin determination. Anal. Chim. Acta 2019, 1077, 255–265. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Canfarotta, F.; Czulak, J.; Johnson, R.; Betlem, K.; Mecozzi, F.; Down, M.P.; Eersels, K.; Van Grinsven, B.; Cleij, T.J.; et al. Thermal Detection of Cardiac Biomarkers Heart-Fatty Acid Binding Protein and ST2 Using a Molecularly Imprinted Nanoparticle-Based Multiplex Sensor Platform. ACS Sensors 2019, 4, 2838–2845. [Google Scholar] [CrossRef]
- Gonçalves, M.D.L.; Truta, L.A.N.; Sales, M.G.F.; Moreira, F.T.C. Electrochemical Point-of Care (PoC) Determination of Interleukin-6 (IL-6) Using a Pyrrole (Py) Molecularly Imprinted Polymer (MIP) on a Carbon-Screen Printed Electrode (C-SPE). Anal. Lett. 2021, 54, 2611–2623. [Google Scholar] [CrossRef]
- Piloto, A.M.L.; Ribeiro, D.S.M.; Rodrigues, S.S.M.; Santos, J.L.M.; Ferreira Sales, M.G. Label-free quantum dot conjugates for human protein IL-2 based on molecularly imprinted polymers. Sensors Actuators B Chem. 2020, 304, 127343. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Zheng, L.; Sun, L.; Jia, W.; Sun, J.; Zhao, X.E.; Liu, H. Derivatization-based magnetic dummy molecularly imprinted polymers integrated with 4-plex stable isotope labeling derivatization strategy for specific and rapid determination of L-hydroxyproline in human serum. Anal. Chim. Acta 2020, 1127, 57–68. [Google Scholar] [CrossRef]
- Arshad, R.; Rhouati, A.; Hayat, A.; Nawaz, M.H.; Yameen, M.A.; Mujahid, A.; Latif, U. MIP-Based Impedimetric Sensor for Detecting Dengue Fever Biomarker. Appl. Biochem. Biotechnol. 2020, 191, 1384–1394. [Google Scholar] [CrossRef]
- Resende, S.; Frasco, M.F.; Sales, M.G.F. A biomimetic photonic crystal sensor for label-free detection of urinary venous thromboembolism biomarker. Sensors Actuators B Chem. 2020, 312, 127947. [Google Scholar] [CrossRef]
- Rebelo, T.S.C.R.; Costa, R.; Brandão, A.T.S.C.; Silva, A.F.; Sales, M.G.F.; Pereira, C.M. Molecularly imprinted polymer SPE sensor for analysis of CA-125 on serum. Anal. Chim. Acta 2019, 1082, 126–135. [Google Scholar] [CrossRef]
- Lanini, S.; Pisapia, R.; Capobianchi, M.R.; Ippolito, G. Global epidemiology of viral hepatitis and national needs for complete control. Expert Rev. Anti. Infect. Ther. 2018, 16, 625–639. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, F.; Chen, C.; Cai, C. Molecular imprinting resonance light scattering nanoprobes based on pH-responsive metal-organic framework for determination of hepatitis A virus. Microchim. Acta 2020, 187, 140. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Shen, X.-L.; Zeng, Q.; Wang, H.-S.; Wang, L.-S. A multi-walled carbon nanotubes based molecularly imprinted polymers electrochemical sensor for the sensitive determination of HIV-p24. Talanta 2017, 164, 121–127. [Google Scholar] [CrossRef]
- Nett, R.J.; Campbell, G.L.; Reisen, W.K. Potential for the Emergence of Japanese Encephalitis Virus in California. Vector-Borne Zoonotic Dis. 2008, 9, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, H.; He, K.; Chen, C.; Chen, X.; Gong, H.; Cai, C. A virus-MIPs fluorescent sensor based on FRET for highly sensitive detection of JEV. Talanta 2016, 160, 360–366. [Google Scholar] [CrossRef]
- Steen Redeker, E.; Eersels, K.; Akkermans, O.; Royakkers, J.; Dyson, S.; Nurekeyeva, K.; Ferrando, B.; Cornelis, P.; Peeters, M.; Wagner, P.; et al. Biomimetic Bacterial Identification Platform Based on Thermal Wave Transport Analysis (TWTA) through Surface-Imprinted Polymers. ACS Infect. Dis. 2017, 3, 388–397. [Google Scholar] [CrossRef] [PubMed]
- van Grinsven, B.; Eersels, K.; Erkens-Hulshof, S.; Diliën, H.; Nurekeyeva, K.; Cornelis, P.; Klein, D.; Crijns, F.; Tuijthof, G.; Wagner, P.; et al. SIP-Based Thermal Detection Platform for the Direct Detection of Bacteria Obtained from a Contaminated Surface. Phys. Status Solidi Appl. Mater. Sci. 2018, 215, 1700777. [Google Scholar] [CrossRef]
- Dasgupta, A. Therapeutic Drug Monitoring; Academic Press: Cambridge, MA, USA, 2012; ISBN 9780123854674. [Google Scholar]
- Alanazi, K.; Garcia Cruz, A.; Di Masi, S.; Voorhaar, A.; Ahmad, O.S.; Cowen, T.; Piletska, E.; Langford, N.; Coats, T.J.; Sims, M.R.; et al. Disposable paracetamol sensor based on electroactive molecularly imprinted polymer nanoparticles for plasma monitoring. Sensors Actuators B Chem. 2021, 329, 129128. [Google Scholar] [CrossRef]
- Yarman, A.; Scheller, F.W. MIP-esterase/Tyrosinase Combinations for Paracetamol and Phenacetin. Electroanalysis 2016, 28, 2222–2227. [Google Scholar] [CrossRef]
- Ozcelikay, G.; Kurbanoglu, S.; Yarman, A.; Scheller, F.W.; Ozkan, S.A. Au-Pt nanoparticles based molecularly imprinted nanosensor for electrochemical detection of the lipopeptide antibiotic drug Daptomycin. Sensors Actuators B Chem. 2020, 320, 128285. [Google Scholar] [CrossRef]
- Liu, H.; Ding, J.; Zhang, K.; Ding, L. Fabrication of carbon dots@restricted access molecularly imprinted polymers for selective detection of metronidazole in serum. Talanta 2020, 209, 120508. [Google Scholar] [CrossRef]
- El-Wekil, M.M.; Mahmoud, A.M.; Marzouk, A.A.; Alkahtani, S.A.; Ali, R. A novel molecularly imprinted sensing platform based on MWCNTs/AuNPs decorated 3D starfish like hollow nickel skeleton as a highly conductive nanocomposite for selective and ultrasensitive analysis of a novel pan-genotypic inhibitor velpatasvir in body fl. J. Mol. Liq. 2018, 271, 105–111. [Google Scholar] [CrossRef]
- Gan, T.; Zhao, A.; Wang, Z.; Liu, P.; Sun, J.; Liu, Y. An electrochemical sensor based on SiO2@TiO2-embedded molecularly imprinted polymers for selective and sensitive determination of theophylline. J. Solid State Electrochem. 2017, 21, 3683–3691. [Google Scholar] [CrossRef]
- Garcia Cruz, A.; Haq, I.; Cowen, T.; Di Masi, S.; Trivedi, S.; Alanazi, K.; Piletska, E.; Mujahid, A.; Piletsky, S.A. Design and fabrication of a smart sensor using in silico epitope mapping and electro-responsive imprinted polymer nanoparticles for determination of insulin levels in human plasma. Biosens. Bioelectron. 2020, 169, 112536. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Cui, F.; Zhuo, B.; Zhao, C.; Liu, W. Sensitive and selective detection of puerarin based on the hybrid of reduced graphene oxide and molecularly imprinted polymer. J. Pharm. Biomed. Anal. 2020, 185, 113221. [Google Scholar] [CrossRef]
- Kamel, A.H.; Amr, A.E.-G.E.; Ashmawy, N.H.; Galal, H.R.; Almehizia, A.A.; Youssef, T.A.; Al-Omar, M.A.; Sayed, A.Y.A. Validation of a novel potentiometric method based on a polymeric PVC membrane sensor integrated with tailored receptors for the antileukemia drug cytarabine. Polymers 2020, 12, 1343. [Google Scholar] [CrossRef] [PubMed]
- Stoian, I.-A.; Iacob, B.-C.; Dudaș, C.-L.; Barbu-Tudoran, L.; Bogdan, D.; Marian, I.O.; Bodoki, E.; Oprean, R. Biomimetic electrochemical sensor for the highly selective detection of azithromycin in biological samples. Biosens. Bioelectron. 2020, 155, 112098. [Google Scholar] [CrossRef]
- Hatamluyi, B.; Hashemzadeh, A.; Darroudi, M. A novel molecularly imprinted polymer decorated by CQDs@HBNNS nanocomposite and UiO-66-NH2 for ultra-selective electrochemical sensing of Oxaliplatin in biological samples. Sensors Actuators B Chem. 2020, 307, 127614. [Google Scholar] [CrossRef]
- Cao, Y.; Feng, T.; Xu, J.; Xue, C. Recent advances of molecularly imprinted polymer-based sensors in the detection of food safety hazard factors. Biosens. Bioelectron. 2019, 141, 111447. [Google Scholar] [CrossRef]
- Peltomaa, R.; Benito-Peña, E.; Moreno-Bondi, M.C. Bioinspired recognition elements for mycotoxin sensors. Anal. Bioanal. Chem. 2018, 410, 747–771. [Google Scholar] [CrossRef]
- Sergeyeva, T.; Yarynka, D.; Piletska, E.; Linnik, R.; Zaporozhets, O.; Brovko, O.; Piletsky, S.; El’skaya, A. Development of a smartphone-based biomimetic sensor for aflatoxin B1 detection using molecularly imprinted polymer membranes. Talanta 2019, 201, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Sergeyeva, T.; Yarynka, D.; Dubey, L.; Dubey, I.; Piletska, E.; Linnik, R.; Antonyuk, M.; Ternovska, T.; Brovko, O.; Piletsky, S.; et al. Sensor based on molecularly imprinted polymer membranes and smartphone for detection of Fusarium contamination in cereals. Sensors 2020, 20, 4304. [Google Scholar] [CrossRef] [PubMed]
- Munawar, H.; Garcia-Cruz, A.; Majewska, M.; Karim, K.; Kutner, W.; Piletsky, S.A. Electrochemical determination of fumonisin B1 using a chemosensor with a recognition unit comprising molecularly imprinted polymer nanoparticles. Sensors Actuators B Chem. 2020, 321, 128552. [Google Scholar] [CrossRef]
- Zhang, W.; Han, Y.; Chen, X.; Luo, X.; Wang, J.; Yue, T.; Li, Z. Surface molecularly imprinted polymer capped Mn-doped ZnS quantum dots as a phosphorescent nanosensor for detecting patulin in apple juice. Food Chem. 2017, 232, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Sequeira, R.; Starbird-Pérez, R.; Rojas-Carillo, O.; Vargas-Villalobos, S. What are the Main Sensor Methods for Quantifying Pesticides in Agricultural Activities? A Review. Molecules 2019, 24, 2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amatatongchai, M.; Thimoonnee, S.; Jarujamrus, P.; Nacapricha, D.; Lieberzeit, P.A. Novel amino-containing molecularly-imprinted polymer coating on magnetite-gold core for sensitive and selective carbofuran detection in food. Microchem. J. 2020, 158, 105298. [Google Scholar] [CrossRef]
- Amatatongchai, M.; Sroysee, W.; Jarujamrus, P.; Nacapricha, D.; Lieberzeit, P.A. Selective amperometric flow-injection analysis of carbofuran using a molecularly-imprinted polymer and gold-coated-magnetite modified carbon nanotube-paste electrode. Talanta 2018, 179, 700–709. [Google Scholar] [CrossRef]
- Cakir, O.; Bakhshpour, M.; Yilmaz, F.; Baysal, Z. Novel QCM and SPR sensors based on molecular imprinting for highly sensitive and selective detection of 2,4-dichlorophenoxyacetic acid in apple samples. Mater. Sci. Eng. C 2019, 102, 483–491. [Google Scholar] [CrossRef]
- Dickert, F.L.; Hayden, O.; Halikias, K.P. Synthetic receptors as sensor coatings for molecules and living cells. Analyst 2001, 126, 766–771. [Google Scholar] [CrossRef]
- Cornelis, P.; Givanoudi, S.; Yongabi, D.; Iken, H.; Duwé, S.; Deschaume, O.; Robbens, J.; Dedecker, P.; Bartic, C.; Wübbenhorst, M.; et al. Sensitive and specific detection of E. coli using biomimetic receptors in combination with a modified heat-transfer method. Biosens. Bioelectron. 2019, 136, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Perçin, I.; Idil, N.; Bakhshpour, M.; Yılmaz, E.; Mattiasson, B.; Denizli, A. Microcontact imprinted plasmonic nanosensors: Powerful tools in the detection of salmonella paratyphi. Sensors 2017, 17, 1375. [Google Scholar] [CrossRef] [Green Version]
- Idil, N.; Hedström, M.; Denizli, A.; Mattiasson, B. Whole cell based microcontact imprinted capacitive biosensor for the detection of Escherichia coli. Biosens. Bioelectron. 2017, 87, 807–815. [Google Scholar] [CrossRef]
- Givanoudi, S.; Cornelis, P.; Rasschaert, G.; Wackers, G.; Iken, H.; Rolka, D.; Yongabi, D.; Robbens, J.; Schöning, M.J.; Heyndrickx, M.; et al. Selective Campylobacter detection and quantification in poultry: A sensor tool for detecting the cause of a common zoonosis at its source. Sensors Actuators B Chem. 2021, 332, 129484. [Google Scholar] [CrossRef]
- Arreguin-Campos, R.; Eersels, K.; Lowdon, J.W.; Rogosic, R.; Heidt, B.; Caldara, M.; Jiménez-Monroy, K.L.; Diliën, H.; Cleij, T.J.; van Grinsven, B. Biomimetic sensing of Escherichia coli at the solid-liquid interface: From surface-imprinted polymer synthesis toward real sample sensing in food safety. Microchem. J. 2021, 169, 106554. [Google Scholar] [CrossRef]
- Ellis, D.I.; Muhamadali, H.; Haughey, S.A.; Elliott, C.T.; Goodacre, R. Point-and-shoot: Rapid quantitative detection methods for on-site food fraud analysis—moving out of the laboratory and into the food supply chain. Anal. Methods 2015, 7, 9401–9414. [Google Scholar] [CrossRef] [Green Version]
- Zeilinger, M.; Sussitz, H.; Cuypers, W.; Jungmann, C.; Lieberzeit, P. Mass-Sensitive Sensing of Melamine in Dairy Products with Molecularly Imprinted Polymers: Matrix Challenges. Sensors 2019, 19, 2366. [Google Scholar] [CrossRef] [Green Version]
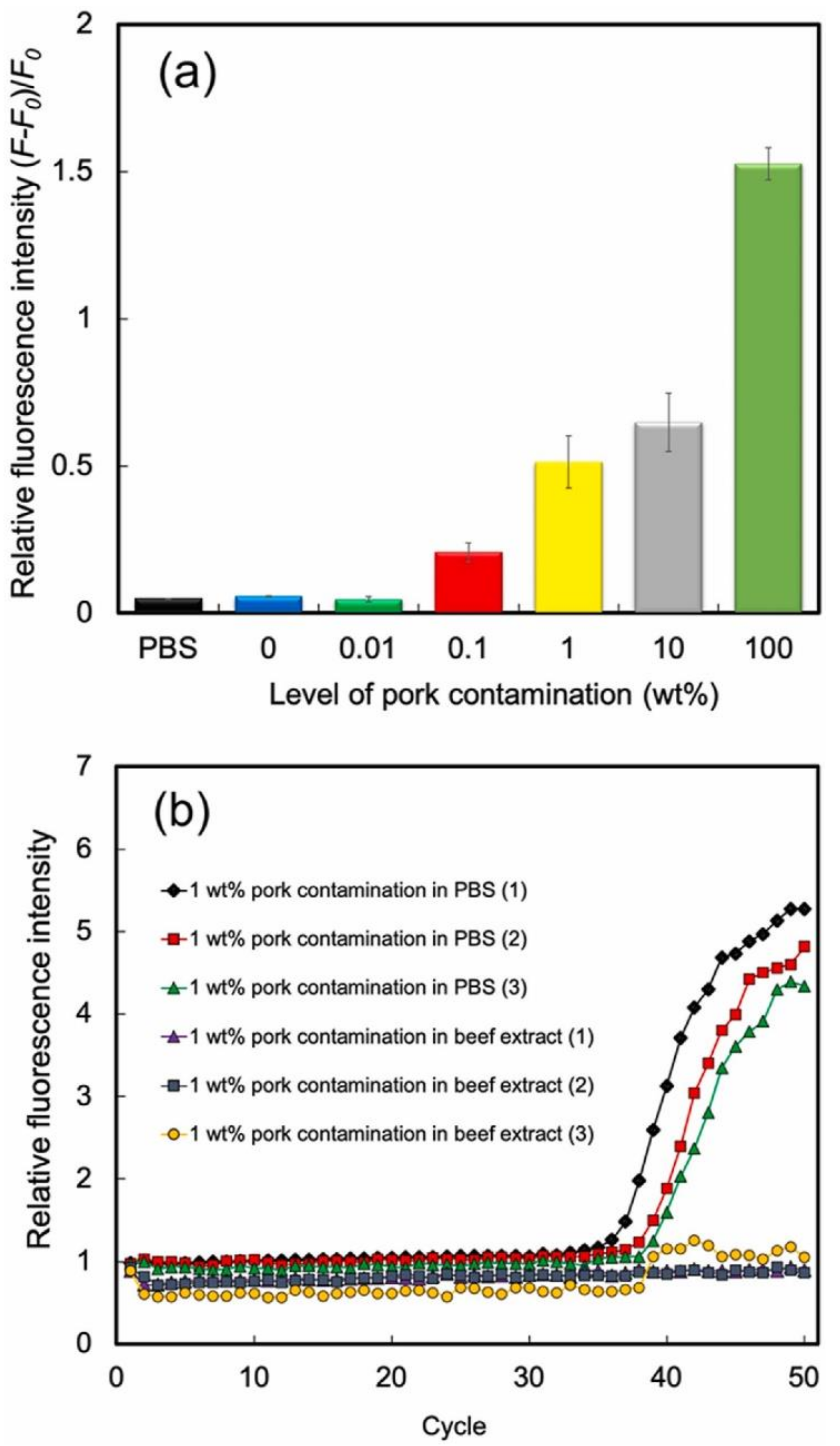
- Cheubong, C.; Takano, E.; Kitayama, Y.; Sunayama, H.; Minamoto, K.; Takeuchi, R.; Furutani, S.; Takeuchi, T. Molecularly imprinted polymer nanogel-based fluorescence sensing of pork contamination in halal meat extracts. Biosens. Bioelectron. 2021, 172, 112775. [Google Scholar] [CrossRef] [PubMed]
- Malitesta, C.; Di Masi, S.; Mazzotta, E. From electrochemical biosensors to biomimetic sensors based on molecularly imprinted polymers in environmental determination of heavy metals. Front. Chem. 2017, 5, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
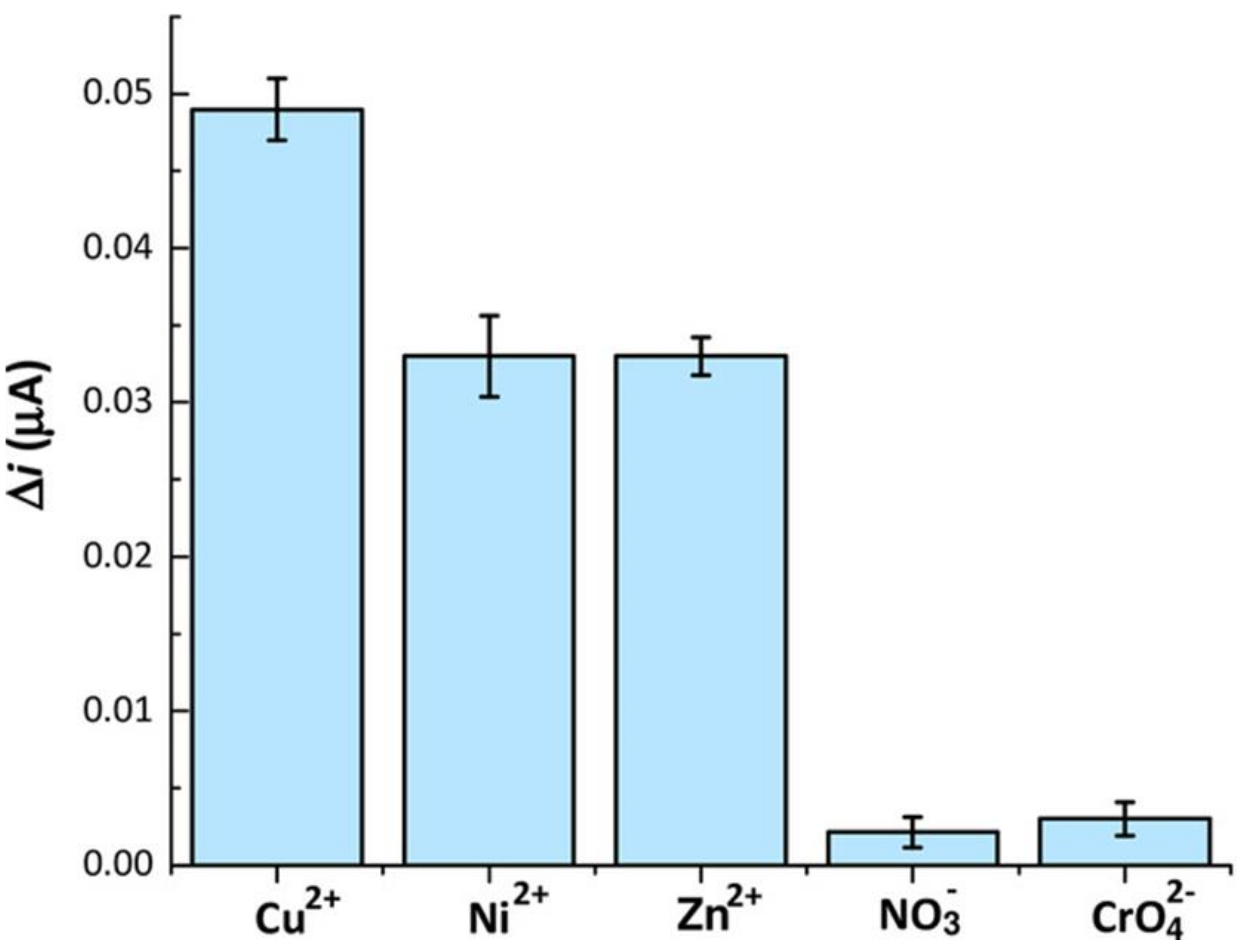
- Di Masi, S.; Garcia Cruz, A.; Canfarotta, F.; Cowen, T.; Marote, P.; Malitesta, C.; Piletsky, S.A. Synthesis and Application of Ion-Imprinted Nanoparticles in Electrochemical Sensors for Copper (II) Determination. ChemNanoMat 2019, 5, 754–760. [Google Scholar] [CrossRef]
- Kumar, S.; Karfa, P.; Patra, S.; Madhuri, R.; Sharma, P.K. Molecularly imprinted star polymer-modified superparamagnetic iron oxide nanoparticle for trace level sensing and separation of mancozeb. RSC Adv. 2016, 6, 36751–36760. [Google Scholar] [CrossRef]
- Farooq, S.; Nie, J.; Cheng, Y.; Yan, Z.; Li, J.; Bacha, S.A.S.; Mushtaq, A.; Zhang, H. Molecularly imprinted polymers’ application in pesticide residue detection. Analyst 2018, 143, 3971–3989. [Google Scholar] [CrossRef]
- El-Akaad, S.; Mohamed, M.A.; Abdelwahab, N.S.; Abdelaleem, E.A.; De Saeger, S.; Beloglazova, N. Capacitive sensor based on molecularly imprinted polymers for detection of the insecticide imidacloprid in water. Sci. Rep. 2020, 10, 14479. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Gálvez, A.; Ait-Lahcen, A.; Mercante, L.A.; Morales-Narváez, E.; Amine, A.; Merkoçi, A. Molecularly Imprinted Polymer-Decorated Magnetite Nanoparticles for Selective Sulfonamide Detection. Anal. Chem. 2016, 88, 3578–3584. [Google Scholar] [CrossRef]
- Hu, Y.; Li, X.; Liu, J.; Wu, M.; Li, M.; Zang, X. One-pot synthesis of a fluorescent molecularly imprinted nanosensor for highly selective detection of sulfapyridine in water. Anal. Methods 2018, 10, 884–890. [Google Scholar] [CrossRef]
- Li, J.; Fu, J.; Yang, Q.; Wang, L.; Wang, X.; Chen, L. Thermosensitive molecularly imprinted core-shell CdTe quantum dots as a ratiometric fluorescence nanosensor for phycocyanin recognition and detection in seawater. Analyst 2018, 143, 3570–3578. [Google Scholar] [CrossRef]
- Sergeyeva, T.; Yarynka, D.; Piletska, E.; Lynnik, R.; Zaporozhets, O.; Brovko, O.; Piletsky, S.; El’skaya, A. Fluorescent sensor systems based on nanostructured polymeric membranes for selective recognition of Aflatoxin B1. Talanta 2017, 175, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Xiao, W.W.; Wang, J.Y.; Xiong, X.H. Rapid isolation and determination of bisphenol A in complicated matrices by magnetic molecularly imprinted electrochemical sensing. Anal. Bioanal. Chem. 2021, 413, 389–401. [Google Scholar] [CrossRef]
- United Nations. World Drug Report 2018; Sales No. E.18.XI.9; UNODC: Vienna, Austria, 2018. [Google Scholar]
- Smolinska-Kempisty, K.; Ahmad, O.S.; Guerreiro, A.; Karim, K.; Piletska, E.; Piletsky, S. New potentiometric sensor based on molecularly imprinted nanoparticles for cocaine detection. Biosens. Bioelectron. 2017, 96, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Grillo, F.; Canfarotta, F.; Whitcombe, M.; Morgan, S.P.; Piletsky, S.; Correia, R.; He, C.Y.; Norris, A.; Korposh, S. Carboxyl-fentanyl detection using optical fibre grating-based sensors functionalised with molecularly imprinted nanoparticles. Biosens. Bioelectron. 2021, 177, 113002. [Google Scholar] [CrossRef] [PubMed]
- De Rycke, E.; Trynda, A.; Jaworowicz, M.; Dubruel, P.; De Saeger, S.; Beloglazova, N. Capacitive sensing of an amphetamine drug precursor in aqueous samples: Application of novel molecularly imprinted polymers for benzyl methyl ketone detection. Biosens. Bioelectron. 2021, 172, 112773. [Google Scholar] [CrossRef]
- Florea, A.; Cowen, T.; Piletsky, S.; De Wael, K. Polymer platforms for selective detection of cocaine in street samples adulterated with levamisole. Talanta 2018, 186, 362–367. [Google Scholar] [CrossRef]
- Florea, A.; Cowen, T.; Piletsky, S.; De Wael, K. Electrochemical sensing of cocaine in real samples based on electrodeposited biomimetic affinity ligands. Analyst 2019, 144, 4639–4646. [Google Scholar] [CrossRef]

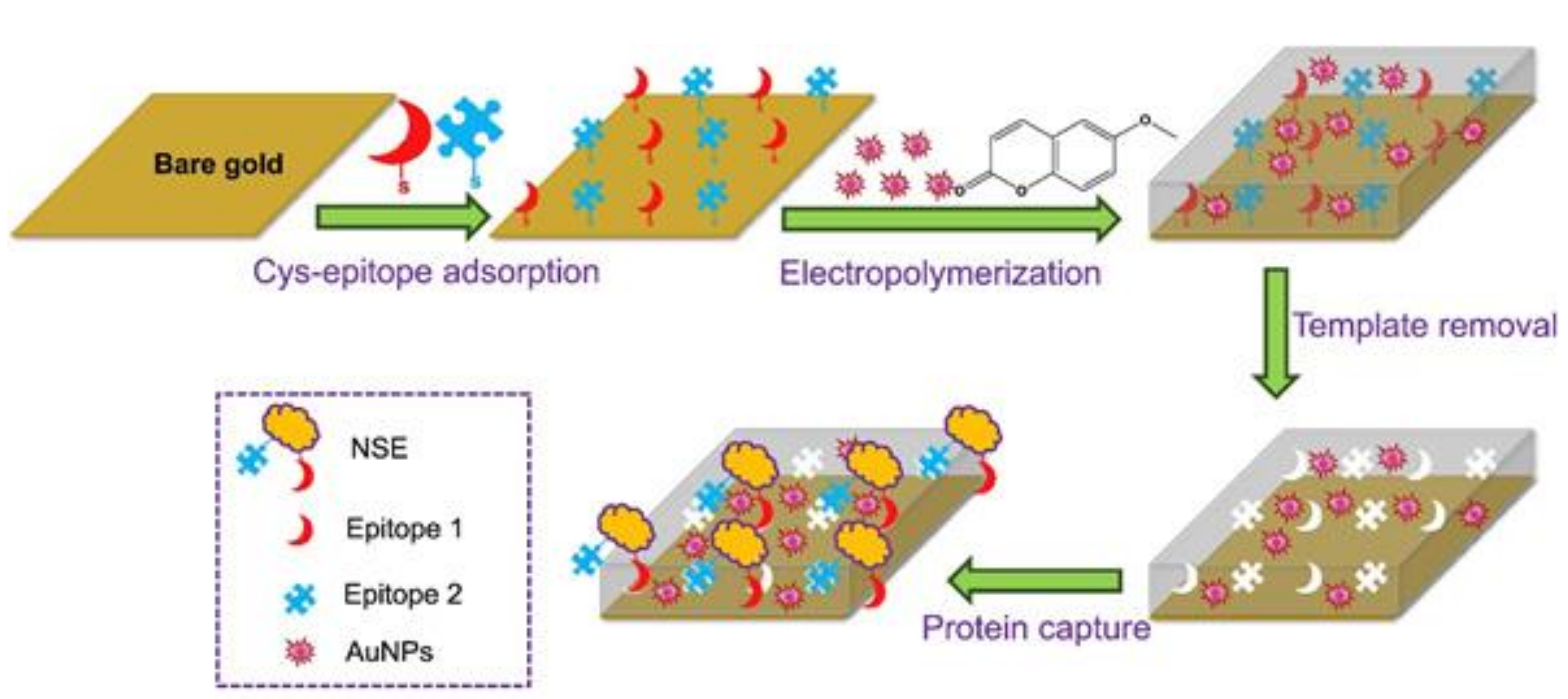
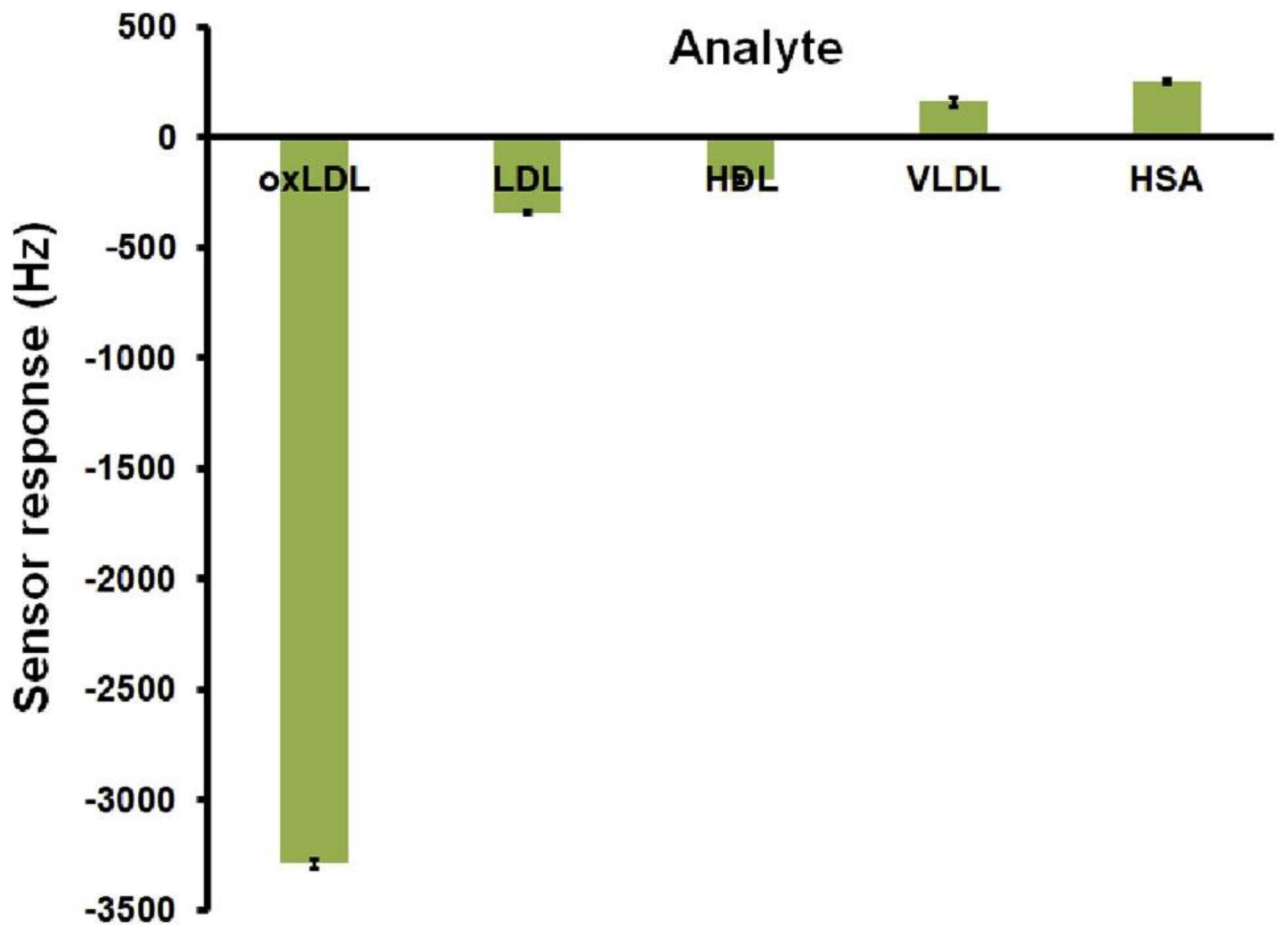
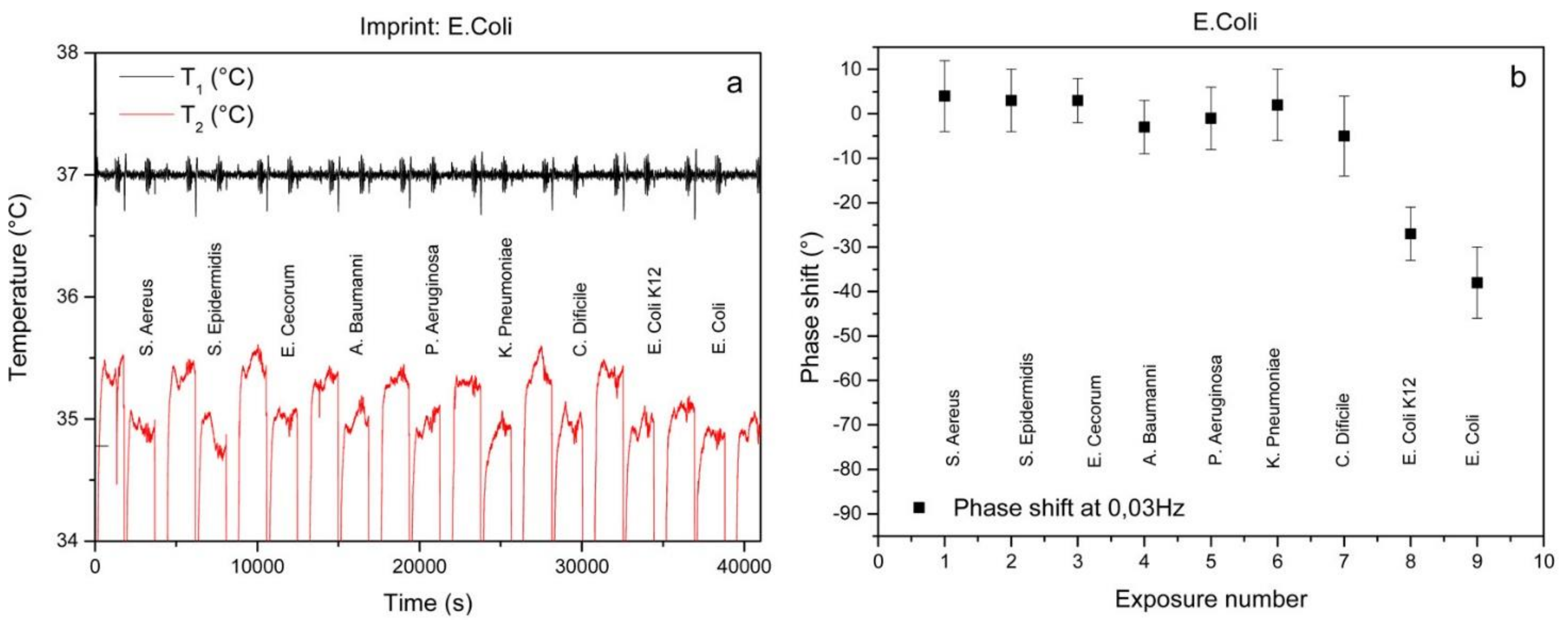
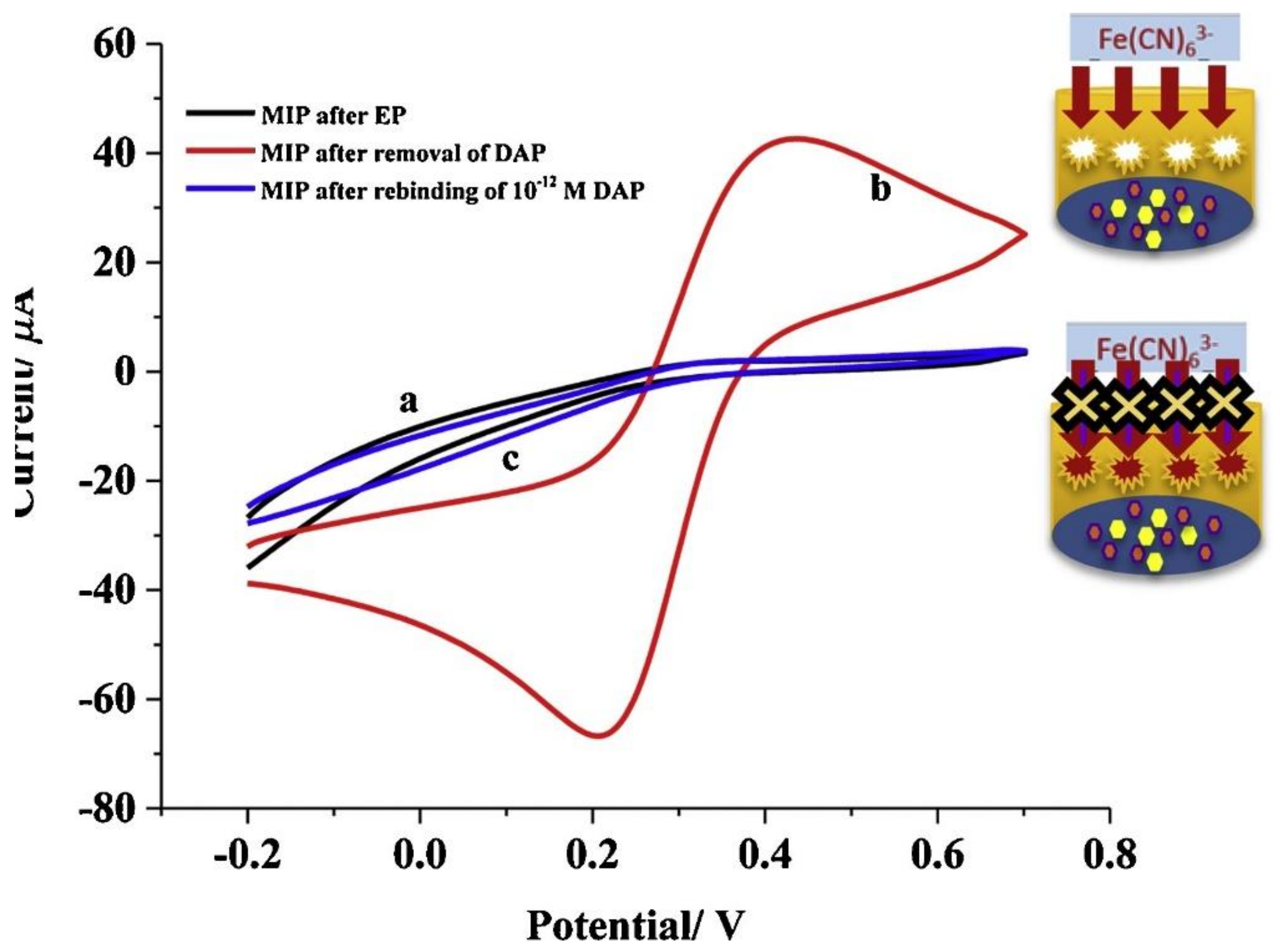



| Biomarker | Linear Range | LOD | Complex Matrix | Recovery Rate [%] | Reference |
|---|---|---|---|---|---|
| Lysozyme | 150 nM–20 µM (PBS) | 141 nM (PBS) | Synthetic saliva | 93–113 | [37] |
| Creatinine | 0.5–150 μM (PBS) | 0.077 μM (PBS) | Human serum and urine | 1.47–5.55% relative difference to reference method | [38] |
| 8-hydroxy-2′-deoxyguanosine | 0.005–50 μM (PBS) | 0.0008 µM (PBS) | Human serum and urine | 94–107 | [38] |
| Serotonin | 0.01–1000 µM (PBS) | 0.002 µM (PBS) | urine | 102–111 | [39] |
| Heart fatty acid binding protein | - | 4.18 ng/mL (PBS) | Fetal bovine serum | - | [40] |
| ST2 | - | 8.79 ng/mL (PBS) | Fetal bovine serum | - | [40] |
| IL-6 | 0.1 pg/mL (PBS) | < 0.1 pg/mL (PBS) | Human serum | - | [41] |
| IL-2 | 35 fg/mL–39 pg/mL (synthetic human serum) | 5.91 fg/mL | Synthetic human serum | - | [42] |
| L-hydroxyproline | 0.2–100 ng/mL (borate buffer) | 0.05 ng/mL (borate buffer) | Human serum | 97.9–102.53 | [43] |
| NS 1 (Dengue Fever biomarker) | 1–200 ng/mL (PBS) | 0.3 ng/mL (PBS) | Human serum | 95–97.14 | [44] |
| Fibrinopeptide B | 0.2–22 ng/mL (urine) | 0.13 ng/mL (urine) | urine | - | [45] |
| CA-125 | 0.01–500 U/mL (PBS) | U/mL (PBS) | Synthetic human serum | 91–105 | [46] |
| Drug | Linear Range | LOD | Complex Matrix | Recovery Rate [%] | Reference |
|---|---|---|---|---|---|
| metronidazole | 50–1200 ng/mL (PBS) | 17.4 ng/mL (PBS) | Human serum | 93.5–102.7 | [58] |
| velpatasvir | 0.649–80.0 ng/mL (phosphate buffer) | 0.21 ng/mL (phosphate buffer) | Plasma and urine | 101.2–101.5 | [59] |
| insulin | 50–2000 pM (plasma) | 0.081 pM | plasma | 99.5–101 | [61] |
| puerarin | 0.02–40 µM | 0.006 µM | urine | 96.7–102.9 | [62] |
| cytarabine | 1.0 × 10−6–1.0 × 10−3 M (phosphate buffer) | 5.5 × 10−7 (phosphate buffer) | Human serum and urine | 98.6–99.1 (urine) 95.3–103 (serum) | [63] |
| azithromycin | 13.33 nM–66.66 µM | 0.85 nM | Plasma, urine and tears | 102.4 (plasma) 98.03–106.33 (urine) 99.06–109.88 (tears) | [64] |
| oxaliplatin | 1.0–20.0 nM 20.0–250.0 nM | 0.39 nM (serum) 0.41 nM (urine) | Human serum and urine | 95.7–106.9 (serum) 94.6–108.3 (urine) | [65] |
| Sample No. | Amount of AFB1 in Sample | Amount of AFB1 Measured with Sensor | Recovery [%] |
|---|---|---|---|
| 1 | 7 µg kg−1 | 6.1 ± 0.52 µg kg−1 | 87 ± 7 |
| 2 | 10 ng mL−1 | 9.6 ± 1.31 ng mL−1 | 96 ± 13 |
| 3 | 50 ng mL−1 | 48.7 ± 1.21 ng mL−1 | 97 ± 2 |
| 4 | 80 ng mL−1 | 76.8 ± 14.69 ng mL−1 | 96 ± 18 |
| 5 | 100 ng mL−1 | 96.3 ± 8.17 ng mL−1 | 96 ± 8 |
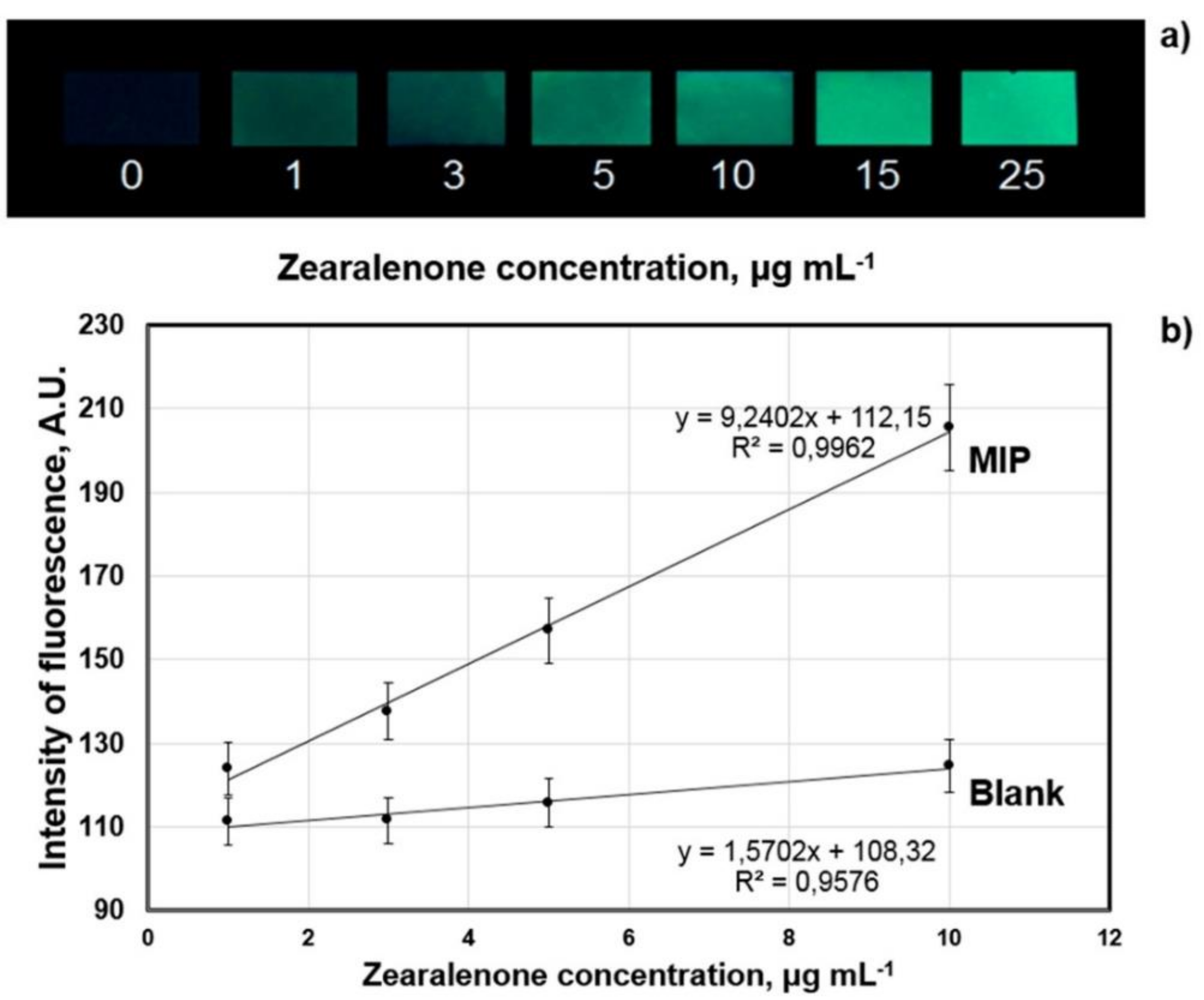
| Sample | Amount of Zearalenone in the Sample | Amount of Zearalenone Determined with Sensor |
|---|---|---|
| maize flour “Dobrodiya Foods”, Kyiv, Ukraine | 1 µg mL−1 | 1.9 ± 0.4 µg mL−1 |
| wheat flour “Kyivmlyn”, Kyiv, Ukraine | 3 µg mL−1 | 4 ± 0.5 µg mL−1 |
| rye flour “Dobrodiya Foods”, Kyiv, Ukraine | 5 µg mL−1 | 5 ± 0.5 µg mL−1 |
| Romer Labs-Check-Sample-Survey CSSMY012-M17161DZ | 114 µg kg−1 | 113.2 ± 7.8 µg kg−1 |
| Detected Bacterium | Complex Matrix | LOD in PBS Buffer [CFUs/mL] | LOD in Matrix [CFUs/mL] | Reference |
|---|---|---|---|---|
| E. coli | 95% apple juice | 100 | 100 | [77] |
| S. paratyphi | apple juice 10× diluted in PBS | 1.4 × 106 | 2.5 × 106 * | [78] |
| E. coli | apple juice 10× diluted in PBS | 70 | 100 * | [79] |
| C. coli | chicken cecal droppings extract in PBS | 1.0 × 103 | 1.1 × 103 | [80] |
| C. jejuni | chicken cecal droppings extract in PBS | 6.6 × 103 | 2.7 × 104 | [80] |
| E. coli | milk | 1.07 × 103 | 2.60 × 103 | [81] |
| No. | AFB1, Added to the Waste Water Sample [ng/mL] | AFB1, Detected in the Waste Water Sample [ng/mL] |
|---|---|---|
| 1 | 50 | 55 ± 6 |
| 2 | 100 | 110 ± 12 |
| 3 | 200 | 285 ± 25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bräuer, B.; Unger, C.; Werner, M.; Lieberzeit, P.A. Biomimetic Sensors to Detect Bioanalytes in Real-Life Samples Using Molecularly Imprinted Polymers: A Review. Sensors 2021, 21, 5550. https://doi.org/10.3390/s21165550
Bräuer B, Unger C, Werner M, Lieberzeit PA. Biomimetic Sensors to Detect Bioanalytes in Real-Life Samples Using Molecularly Imprinted Polymers: A Review. Sensors. 2021; 21(16):5550. https://doi.org/10.3390/s21165550
Chicago/Turabian StyleBräuer, Birgit, Christine Unger, Martin Werner, and Peter A. Lieberzeit. 2021. "Biomimetic Sensors to Detect Bioanalytes in Real-Life Samples Using Molecularly Imprinted Polymers: A Review" Sensors 21, no. 16: 5550. https://doi.org/10.3390/s21165550
APA StyleBräuer, B., Unger, C., Werner, M., & Lieberzeit, P. A. (2021). Biomimetic Sensors to Detect Bioanalytes in Real-Life Samples Using Molecularly Imprinted Polymers: A Review. Sensors, 21(16), 5550. https://doi.org/10.3390/s21165550






