Hyperglycemia Identification Using ECG in Deep Learning Era
Abstract
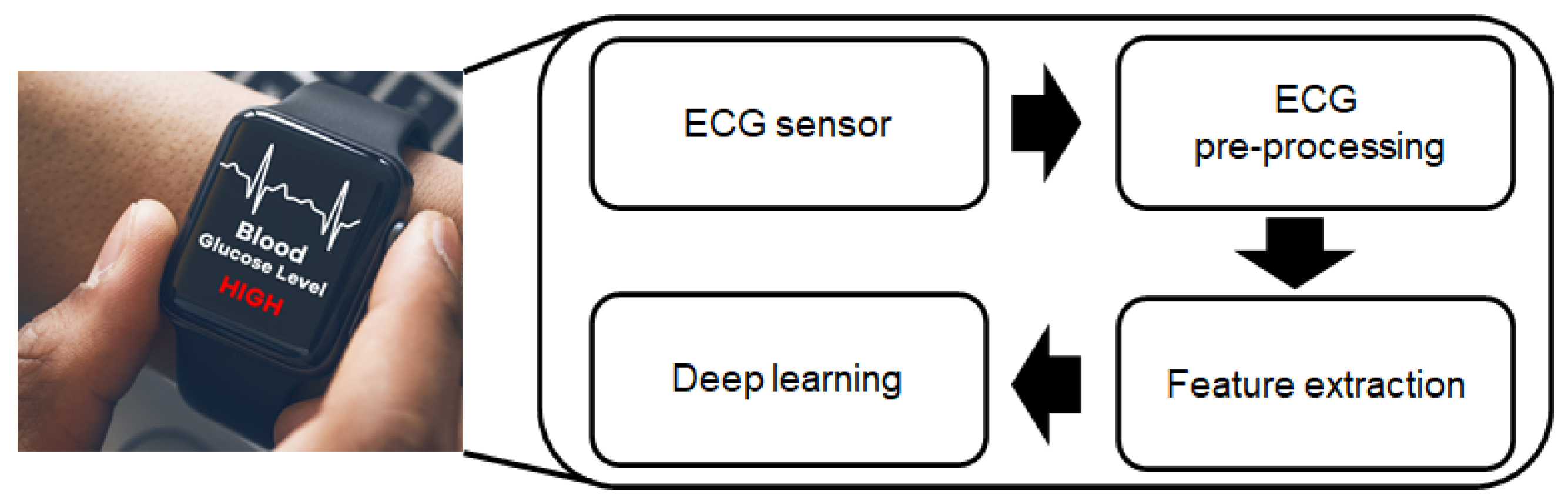
:1. Introduction
- We develop a 10-layer deep learning based hyperglycemia detection technique and more robust approaches for processing ECGs.
- We present different feature extraction techniques. Specifically, we investigate novel fiducial methods such as slope and temporal and amplitude characteristics. This resulted in a feature size reduction of 97% when compared to a full ECG cardiac cycle.
- To demonstrate the effectiveness, robustness, and generalization ability of our proposed methods, we conducted experiments on a new ECG database containing 68,274 samples collected from 1119 subjects.
- We provide detailed classification analysis of age, weight, height, and heart rate and discuss the impact of these on hyperglycemia.
2. Background
2.1. Hyperglycemia
2.2. Electrocardiogram (ECG)
3. Literature Review
4. The Proposed Approach
4.1. Filtering
4.2. Fiducial Points and Cardiac Cycles Identification
4.3. Features Extraction
4.4. QT Correction
4.5. Outliers Removal
4.6. Normalization
5. Experimental Setup
5.1. Dataset
- Each subject participated in two sequential recording sessions, both taken in the morning.
- Each session consisted of the recording of a 60-s single-lead ECG and blood glucose concentration.
- ECG was acquired using Analog AD-8232 with a sampling rate of 1000 Hz [46].
- Blood glucose concentration was measured using Accu-Chek Mobile blood glucose monitoring system [47].
5.2. Hardware and Software
5.3. Training and Testing
5.4. Models and Metrics
6. Experimental Result
Results Discussion
7. Challenge
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Detailed Models Simulations Results
| C or Degree | Logistic Regression | SVM Linear | SVM Gaussian | SVM Polynomial |
|---|---|---|---|---|
| 0.001 | 61.45% | 58.02% | 18.60% | - |
| 0.01 | 62.09% | 53.86% | 18.16% | - |
| 0.1 | 61.87% | 42.17% | 17.61% | - |
| 1 | 62.37% | 57.36% | 47.90% | 55.92% |
| 2 | 61.87% | 44.01% | 52.03% | 55.14% |
| 3 | 61.87% | 57.57% | 52.03% | 42.74% |
| 4 | 61.92% | 43.47% | 52.03% | 48.17% |
| 5 | 62.44% | 42.22% | 52.03% | 41.85% |
| 6 | 61.95% | 41.88% | 52.03% | 56.36% |
| 7 | 61.91% | 41.96% | 52.03% | 50.15% |
| 8 | 62.43% | 44.88% | 52.03% | 50.00% |
| 9 | 61.85% | 50.16% | 52.03% | 50.00% |
| 10 | 62.39% | 41.37% | 52.03% | 50.00% |
| 20 | 61.86% | 57.93% | 52.03% | 50.00% |
| 30 | 61.84% | 48.64% | 52.03% | 50.00% |
| 40 | 61.90% | 41.66% | 52.03% | 50.00% |
| 50 | 62.44% | 58.99% | 52.03% | 50.00% |
| 60 | 61.86% | 57.56% | 52.03% | 50.00% |
| 70 | 61.87% | 56.22% | 52.03% | 50.00% |
| 80 | 61.93% | 51.63% | 52.03% | 50.00% |
| 90 | 61.92% | 56.93% | 52.03% | 50.00% |
| 100 | 62.37% | 42.46% | 52.03% | 50.00% |
| # of Units per Layer (exc. Output Layer) | 100 | 200 | 300 | 400 | 500 | |
|---|---|---|---|---|---|---|
| # of Layers | ||||||
| 2 | 49.96% | 50.00% | 50.00% | 79.64% | 50.00% | |
| 3 | 79.34% | 50.00% | 50.00% | 50.00% | 50.00% | |
| 4 | 80.46% | 49.99% | 88.68% | 50.00% | 89.25% | |
| 5 | 83.64% | 87.02% | 88.57% | 90.76% | 50.00% | |
| 6 | 82.94% | 89.85% | 91.68% | 91.06% | 92.53% | |
| 7 | 85.69% | 90.78% | 91.94% | 92.43% | 93.20% | |
| 8 | 86.57% | 89.49% | 92.76% | 92.26% | 93.44% | |
| 9 | 88.81% | 89.96% | 92.80% | 92.91% | 93.59% | |
| 10 | 89.06% | 91.88% | 92.29% | 94.34% | 94.53% |
References
- Centers for Disease Control (CDC). Available online: https://www.cdc.gov/heartdisease/facts.htm (accessed on 30 July 2021).
- Wang, P.; Hou, B.; Shao, S.; Yan, R. ECG Arrhythmias Detection Using Auxiliary Classifier Generative Adversarial Network and Residual Network. IEEE Access 2019, 7, 100910–100922. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans. Biomed. Eng. 2015, 63, 664–675. [Google Scholar] [CrossRef]
- Sun, L.; Lu, Y.; Yang, K.; Li, S. ECG analysis using multiple instance learning for myocardial infarction detection. IEEE Trans. Biomed. Eng. 2012, 59, 3348–3356. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, Y.; He, J.; Wang, L.; Tian, Y.; Zhou, T.s.; Li, T.; Li, J.s. High-performance personalized heartbeat classification model for long-term ECG signal. IEEE Trans. Biomed. Eng. 2016, 64, 78–86. [Google Scholar]
- Nguyen, L.L.; Su, S.; Nguyen, H.T. Neural network approach for non-invasive detection of hyperglycemia using electrocardiographic signals. In Proceedings of the 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2014; pp. 4475–4478. [Google Scholar] [CrossRef]
- Leal, Y.; Gonzalez-Abril, L.; Lorencio, C.; Bondia, J.; Vehi, J. Detection of correct and incorrect measurements in real-time continuous glucose monitoring systems by applying a postprocessing support vector machine. IEEE Trans. Biomed. Eng. 2013, 60, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Turksoy, K.; Roy, A.; Cinar, A. Real-Time Model-Based Fault Detection of Continuous Glucose Sensor Measurements. IEEE Trans. Biomed. Eng. 2017, 64, 1437–1445. [Google Scholar] [CrossRef] [PubMed]
- Diabetes. Available online: https://www.who.int/news-room/fact-sheets/detail/diabetes (accessed on 30 July 2021).
- Statistics about Diabetes. Available online: https://www.diabetes.org/resources/statistics/statistics-about-diabetes (accessed on 30 July 2021).
- Caramelo, F.; Ferreira, N.; Oliveiros, B. Estimation of risk factors for COVID-19 mortality-preliminary results. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Ma, P.; Zhang, S.; Song, S.; Wang, Z.; Ma, Y.; Xu, J.; Wu, F.; Duan, L.; Yin, Z.; et al. Fasting blood glucose at admission is an independent predictor for 28-day mortality in patients with COVID-19 without previous diagnosis of diabetes: A multi-centre retrospective study. Diabetologia 2020, 63, 2102–2111. [Google Scholar] [CrossRef]
- Taubes, G.; Chamberlain, M. Why We Get Fat; Joosr Ltd.: Hove, UK, 2016. [Google Scholar]
- Vedanthan, R.; Fuster, V.; Fischer, A. Sudden cardiac death in low-and middle-income countries. Glob. Heart 2012, 7, 353–360. [Google Scholar] [CrossRef] [Green Version]
- Taylor, R. Insulin resistance and type 2 diabetes. Diabetes 2012, 61, 778–779. [Google Scholar] [CrossRef] [Green Version]
- U.S. Food and Drug Administration. Self-Monitoring Blood Glucose Test Systems for Over-the-Counter Use—Guidance for Industry and Food and Drug Administration Staff; U.S. Food and Drug Administration: Rockville, MD, USA, 2016.
- Thompson, N.D.; Perz, J.F. Eliminating the Blood: Ongoing Outbreaks of Hepatitis B Virus Infection and the Need for Innovative Glucose Monitoring Technologies. J. Diabetes Sci. Technol. 2009, 3, 283–288. [Google Scholar] [CrossRef] [Green Version]
- Ali, H.; Bensaali, F.; Jaber, F. Novel Approach to Non-Invasive Blood Glucose Monitoring Based on Transmittance and Refraction of Visible Laser Light. IEEE Access 2017, 5, 9163–9174. [Google Scholar] [CrossRef]
- Malmivuo, P.; Malmivuo, J.; Plonsey, R. Bioelectromagnetism: Principles and Applications of Bioelectric and Biomagnetic Fields; Oxford University Press: New York, NY, USA, 1995. [Google Scholar]
- Kost, J.; Mitragotri, S.; Gabbay, R.A.; Pishko, M.; Langer, R. Transdermal monitoring of glucose and other analytes using ultrasound. Nat. Med. 2000, 6, 347. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.S.; Jyostna, K. Development of Smart Insulin Device for Non Invasive Blood Glucose Level Monitoring. In Proceedings of the IEEE 7th International Advance Computing Conference (IACC), Hyderabad, India, 5–7 January 2017; pp. 516–519. [Google Scholar] [CrossRef]
- Julian, E.S.; Prawiroredjo, K.; Tjahjadi, G. The Model of near infrared sensor output voltage as a function of glucose concentration in solution. In Proceedings of the 15th International Conference on Quality in Research (QiR): International Symposium on Electrical and Computer Engineering, Nusa Dua, Bali, Indonesia, 24–27 July 2017; pp. 146–149. [Google Scholar] [CrossRef]
- Pai, P.P.; Sanki, P.K.; Sahoo, S.K.; De, A.; Bhattacharya, S.; Banerjee, S. Cloud Computing-Based Non-Invasive Glucose Monitoring for Diabetic Care. IEEE Trans. Circuits Syst. I Regul. Pap. 2018, 65, 663–676. [Google Scholar] [CrossRef]
- Anas, M.N.; Nurun, N.K.; Norali, A.N.; Normahira, M. Non-invasive blood glucose measurement. In Proceedings of the 2012 IEEE-EMBS Conference on Biomedical Engineering and Sciences, Langkawi, Malaysia, 17–19 December 2012; pp. 503–507. [Google Scholar] [CrossRef]
- Liu, Y.; Xia, M.; Nie, Z.; Li, J.; Zeng, Y.; Wang, L. In vivo wearable non-invasive glucose monitoring based on dielectric spectroscopy. In Proceedings of the IEEE 13th International Conference on Signal Processing (ICSP), Chengdu, China, 6–10 November 2016; pp. 1388–1391. [Google Scholar] [CrossRef]
- Vilaboy, M.J.; Ergin, A.; Tchouassi, A.; Greene, R.; Thomas, G.A. Optical measurement of glucose concentrations using Raman spectroscopy. In Proceedings of the 2003 IEEE 29th Annual Proceedings of Bioengineering Conference; Newark, NJ, USA, 22–23 March 2003, pp. 329–330. [CrossRef]
- Amanipour, R.; Nazeran, H.; Reyes, I.; Franco, M.; Haltiwanger, E. The effects of blood glucose changes on frequency-domain measures of HRV signal in type 1 diabetes. In Proceedings of the CONIELECOMP 2012, 22nd International Conference on Electrical Communications and Computers, Puebla, Mexico, 27–29 February 2012; pp. 50–54. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Fukuki, M.; Hoshio, A.; Sasaki, N.; Hamada, T.; Tanaka, Y.; Yoshida, A.; Shigemasa, C.; Mashiba, H. Decreased heart rate variability in patients with diabetes mellitus and ischemic heart disease. Jpn. Circ. J. 1996, 60, 925–932. [Google Scholar] [CrossRef] [Green Version]
- Perpiñan, G.; Severeyn, E.; Wong, S.; Altuve, M. Nonlinear heart rate variability measures during the oral glucose tolerance test. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Farina, P.V.R.; Severeyn, E.; Wong, S.; Turiel, J.P. Study of cardiac repolarization during Oral Glucose Tolerance Test in metabolic syndrome patients. In Proceedings of the 2012 Computing in Cardiology, Krakow, Poland, 9–12 September 2012; pp. 429–432. [Google Scholar]
- Farina, P.V.R.; Pérez Turiel, J.; Pagán-Buzo, F.J.; González Sarmiento, E.; Herreros López, A.; Rodríguez-Guerrero, C.D. QTc analysis and comparison in pre-diabetic patients. In Proceedings of the 2010 Computing in Cardiology, Belfast, UK, 26–29 September 2010; pp. 697–700. [Google Scholar]
- Suys, B.; Heuten, S.; De Wolf, D.; Verherstraeten, M.; de Beeck, L.O.; Matthys, D.; Vrints, C.; Rooman, R. Glycemia and corrected QT interval prolongation in young type 1 diabetic patients: What is the relation? Diabetes Care 2006, 29, 427–429. [Google Scholar] [CrossRef] [Green Version]
- Marfella, R.; Nappo, F.; De Angelis, L.; Siniscalchi, M.; Rossi, F.; Giugliano, D. The effect of acute hyperglycaemia on QTc duration in healthy man. Diabetologia 2000, 43, 571–575. [Google Scholar] [CrossRef]
- Nguyen, L.L.; Su, S.; Nguyen, H.T. Identification of Hypoglycemia and Hyperglycemia in Type 1 Diabetic patients using ECG parameters. In Proceedings of the 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; pp. 2716–2719. [Google Scholar] [CrossRef]
- Task Force of the European Society of Cardiology the North American Society of Pacing Electrophysiology. Heart rate variability: Standards of measurement, physiological interpretation, and clinical use. Circulation 1996, 93, 1043–1065. [Google Scholar] [CrossRef] [Green Version]
- Ingale, M.; Cordeiro, R.; Thentu, S.; Park, Y.; Karimian, N. Ecg biometric authentication: A comparative analysis. IEEE Access 2020, 8, 117853–117866. [Google Scholar] [CrossRef]
- Carreiras, C.; Alves, A.P.; Lourenço, A.; Canento, F.; Silva, H.; Fred, A. BioSPPy: Biosignal Processing in Python. Available online: https://github.com/PIA-Group/BioSPPy (accessed on 30 July 2021).
- Hamilton, P.S.; Tompkins, W.J. Quantitative Investigation of QRS Detection Rules Using the MIT/BIH Arrhythmia Database. IEEE Trans. Biomed. Eng. 1986, BME-33, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Makowski, D. NeuroKit: A Python Toolbox for Statistics and Neurophysiological Signal Processing (EEG, EDA, ECG, EMG…). Available online: https://github.com/neuropsychology/NeuroKit.p (accessed on 30 July 2021).
- Karimian, N.; Guo, Z.; Tehranipoor, M.; Forte, D. Highly Reliable Key Generation From Electrocardiogram (ECG). IEEE Trans. Biomed. Eng. 2017, 64, 1400–1411. [Google Scholar] [CrossRef]
- Malik, M. Problems of heart rate correction in assessment of drug-induced QT interval prolongation. J. Cardiovasc. Electrophysiol. 2001, 12, 411–420. [Google Scholar] [CrossRef]
- Vandenberk, B.; Vandael, E.; Robyns, T.; Vandenberghe, J.; Garweg, C.; Foulon, V.; Ector, J.; Willems, R. Which QT correction formulae to use for QT monitoring? J. Am. Heart Assoc. 2016, 5, e003264. [Google Scholar] [CrossRef] [PubMed]
- Rousseeuw, P.J.; Hubert, M. Robust statistics for outlier detection. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011, 1, 73–79. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AD8232 Datasheet and Product Info|Analog Devices. Available online: https://www.analog.com/en/products/ad8232.html#product-overview (accessed on 30 July 2021).
- The Accu-Chek Mobile System|Accu-Chek. Available online: https://www.accu-chek.co.uk/blood-glucose-meters/mobile#product-specs (accessed on 30 July 2021).
- Tarvainen, M.P.; Niskanen, J.P.; Lipponen, J.A.; Ranta-Aho, P.O.; Karjalainen, P.A. Kubios HRV–heart rate variability analysis software. Comput. Methods Programs Biomed. 2014, 113, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Swapna, G.; Vinayakumar, R.; Soman, K. Diabetes detection using deep learning algorithms. ICT Express 2018, 4, 243–246. [Google Scholar]
- Singh, J.P.; Larson, M.G.; O’Donnell, C.J.; Wilson, P.F.; Tsuji, H.; Lloyd-Jones, D.M.; Levy, D. Association of hyperglycemia with reduced heart rate variability (The Framingham Heart Study). Am. J. Cardiol. 2000, 86, 309–312. [Google Scholar] [CrossRef]
- Faust, O.; Acharya, U.R.; Molinari, F.; Chattopadhyay, S.; Tamura, T. Linear and non-linear analysis of cardiac health in diabetic subjects. Biomed. Signal Process. Control 2012, 7, 295–302. [Google Scholar] [CrossRef]
- Li, J.; Igbe, T.; Liu, Y.; Kandwal, A.; Wang, L.; Nie, Z. Non-invasive monitoring of three glucose ranges based on ECG by using DBSCAN-CNN. IEEE J. Biomed. Health Inform. 2021, 25, 3340–3350. [Google Scholar] [CrossRef] [PubMed]







| # | Feature | Type |
|---|---|---|
| 1 | HR | Intervals |
| 2 | PR | Intervals |
| 3 | QTc | Intervals |
| 4 | RTc | Intervals |
| 5 | TpTec | Intervals |
| 6 | Mean RR interval | Time-domain |
| 7 | Standard deviation of the RR Interval index (SDNN) | Time-domain |
| 8 | Root mean square of successive RR interval differences (RMSSD) | Time-domain |
| 9 | Percentage of consecutive RR intervals that differ by more than 50 ms (pNN50) | Time-domain |
| 10 | HRV triangular index (HRVi) | Time-domain |
| 11 | Baseline width of the RR interval histogram evaluated through triangular interpolation (TINN) | Time-domain |
| 12 | Very low frequency (VLF) | Frequency-domain |
| 13 | Low frequency (LF) | Frequency-domain |
| 14 | High frequency (HF) | Frequency-domain |
| 15 | Total spectral power (TotalPw) | Frequency-domain |
| 16 | LF/HF ratio | Frequency-domain |
| # | Feature | # | Feature |
|---|---|---|---|
| 1 | PQ length | 10 | QR slope |
| 2 | PQ slope | 11 | QS length |
| 3 | PR length | 12 | QS slope |
| 4 | PR slope | 13 | QT length |
| 5 | PS length | 14 | QT slope |
| 6 | PS slope | 15 | RS length |
| 7 | PT length | 16 | RS slope |
| 8 | PT slope | 17 | RT length |
| 9 | QR length | 18 | RT slope |
| Model | AUC |
|---|---|
| 10-layer DNN | 94.53% |
| Logistic Regression (C = 5) | 62.44% |
| SVM Linear (C = 50) | 58.99% |
| SVM Polynomial (d = 6) | 56.36% |
| SVM Gaussian (C = 2) | 52.03% |
| k | AUC | k | AUC |
|---|---|---|---|
| 1 | 96.98% | 6 | 97.17% |
| 2 | 97.23% | 7 | 97.43% |
| 3 | 96.40% | 8 | 98.23% |
| 4 | 97.34% | 9 | 96.94% |
| 5 | 96.03% | 10 | 95.49% |
| Sensitivity | Specificity | AUC | |
|---|---|---|---|
| 10-layer DNN | 87.57% | 85.04% | 94.53% |
| 3-layer ANN [6] modified | 65.64% | 56.21% | 61.68% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cordeiro, R.; Karimian, N.; Park, Y. Hyperglycemia Identification Using ECG in Deep Learning Era. Sensors 2021, 21, 6263. https://doi.org/10.3390/s21186263
Cordeiro R, Karimian N, Park Y. Hyperglycemia Identification Using ECG in Deep Learning Era. Sensors. 2021; 21(18):6263. https://doi.org/10.3390/s21186263
Chicago/Turabian StyleCordeiro, Renato, Nima Karimian, and Younghee Park. 2021. "Hyperglycemia Identification Using ECG in Deep Learning Era" Sensors 21, no. 18: 6263. https://doi.org/10.3390/s21186263
APA StyleCordeiro, R., Karimian, N., & Park, Y. (2021). Hyperglycemia Identification Using ECG in Deep Learning Era. Sensors, 21(18), 6263. https://doi.org/10.3390/s21186263








