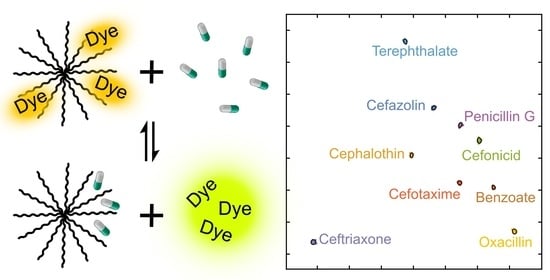
Beta-Lactam Antibiotic Discrimination Using a Macromolecular Sensor in Water at Neutral pH
Abstract
:1. Introduction
2. Materials and Methods
3. Results
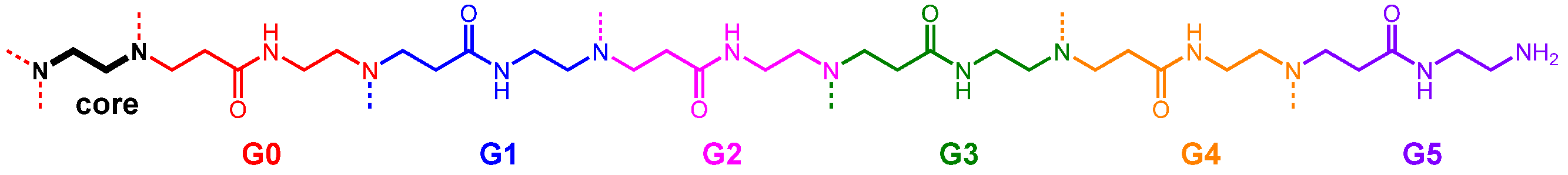
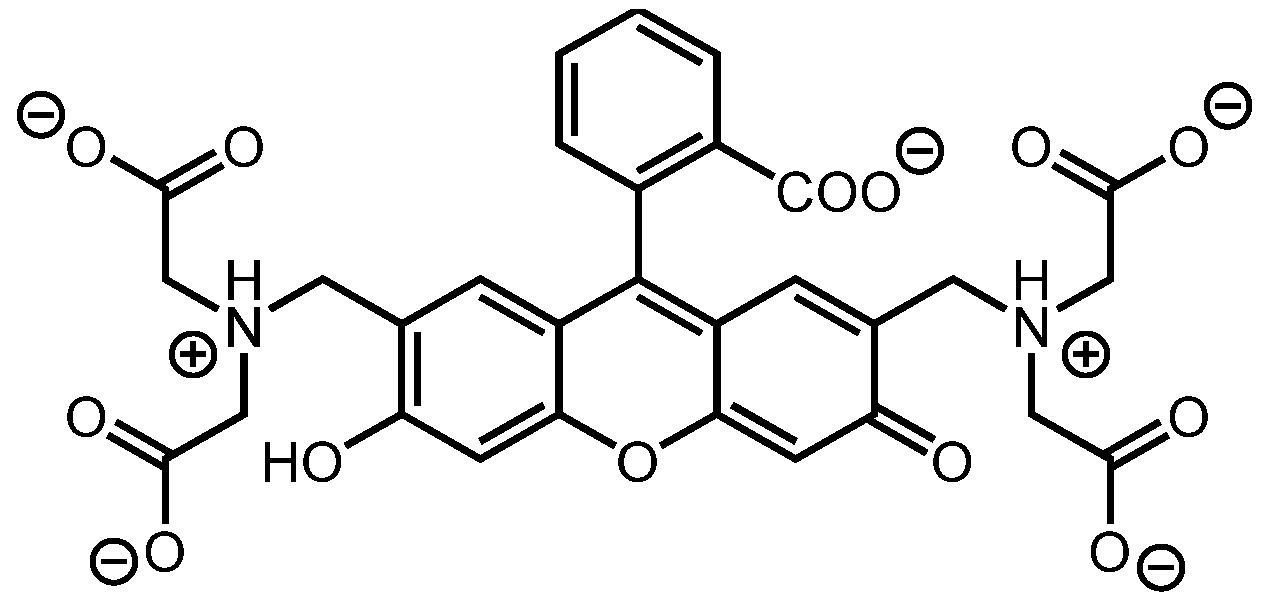
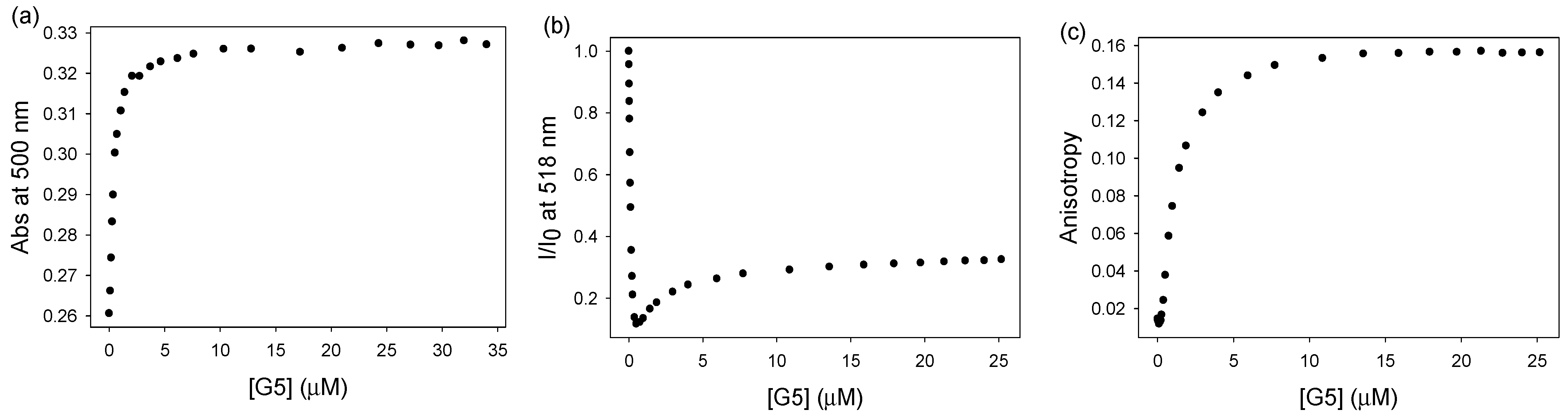
3.1. Indicator Displacement Setup: Binding of Calcein Dye to PAMAM G5 Dendrimer
3.2. Binding of Carboxylate to PAMAM G5 Dendrimer (Indicator Displacement Assay)
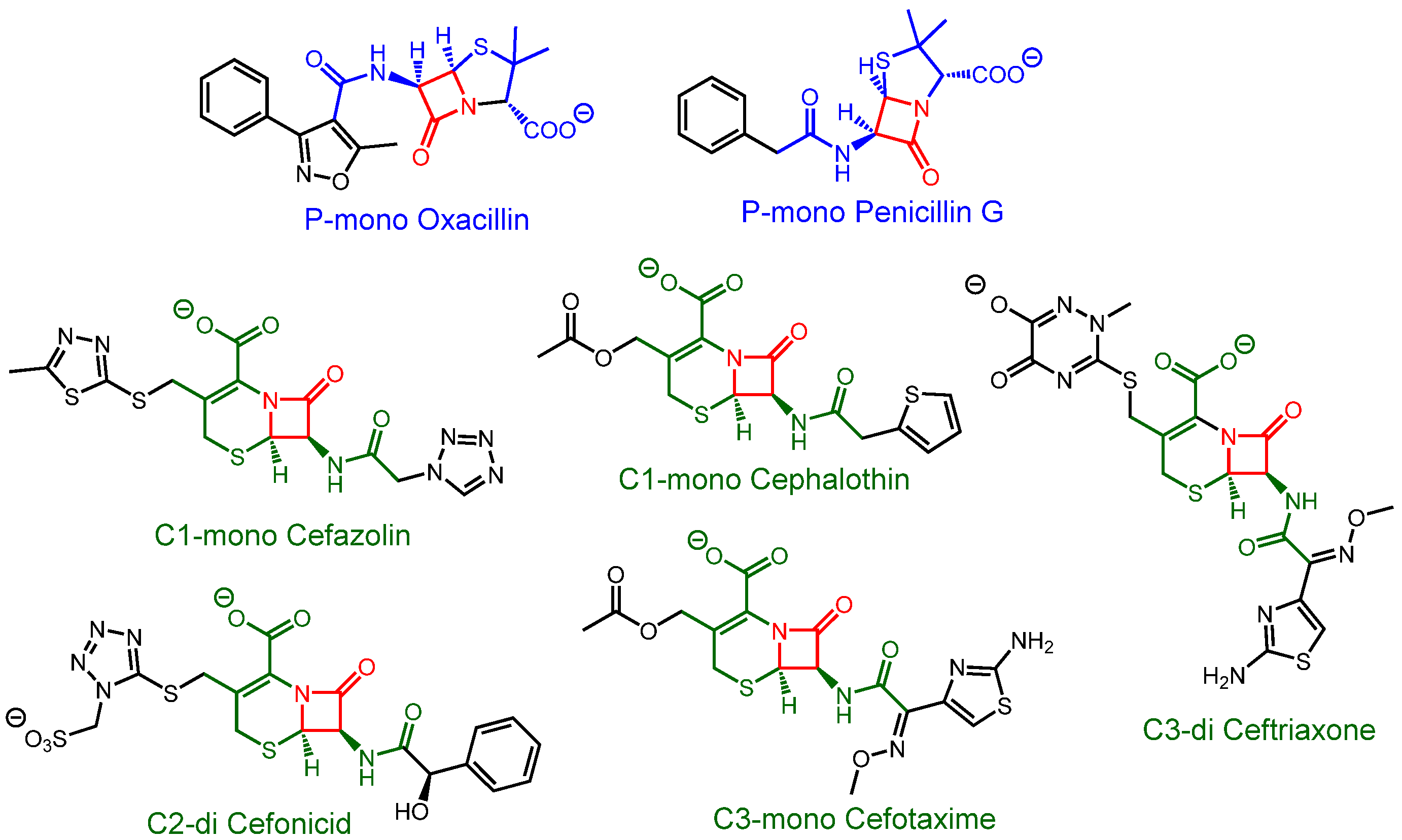
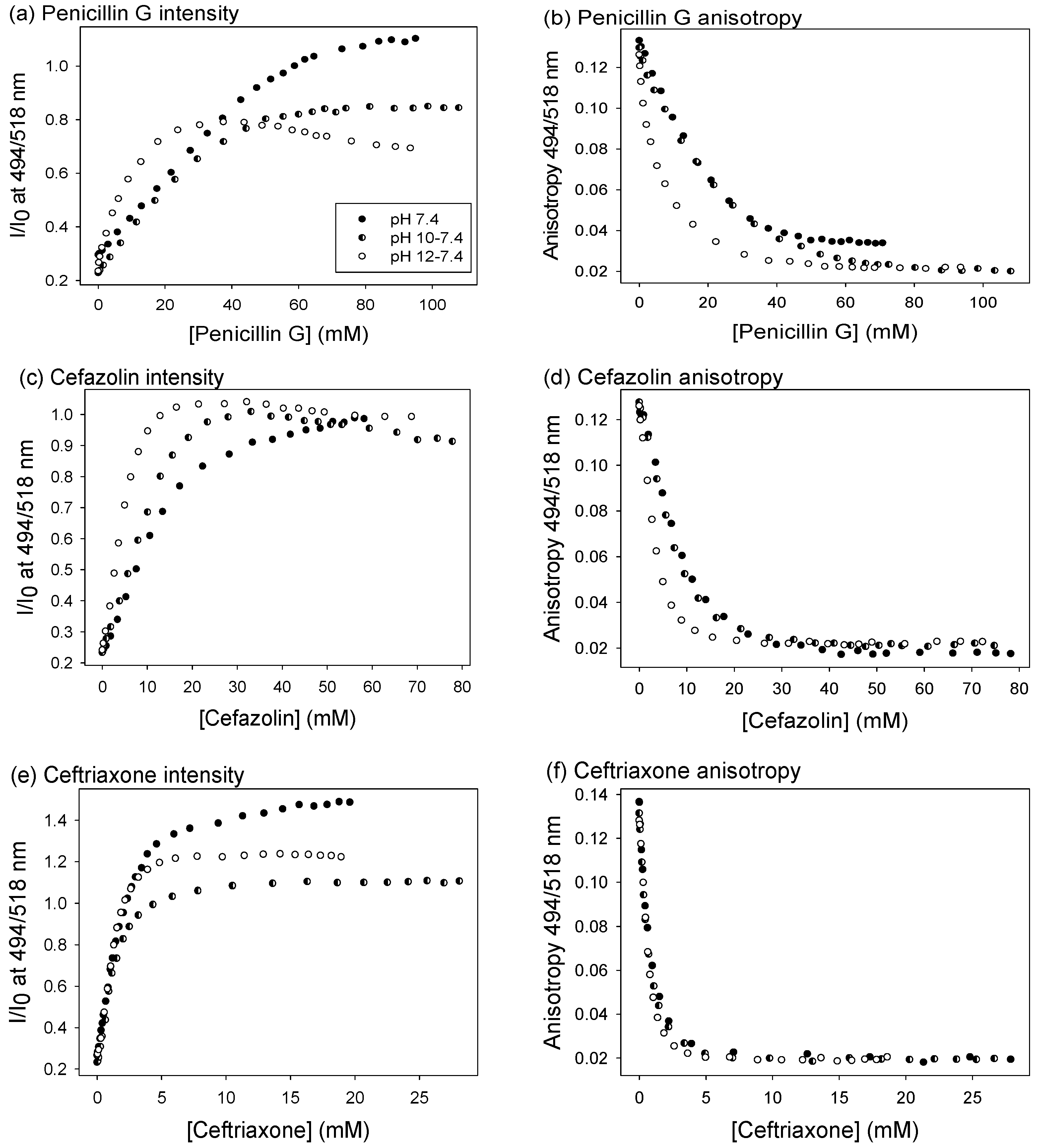
3.3. Antibiotics Discrimination at pH 7.4
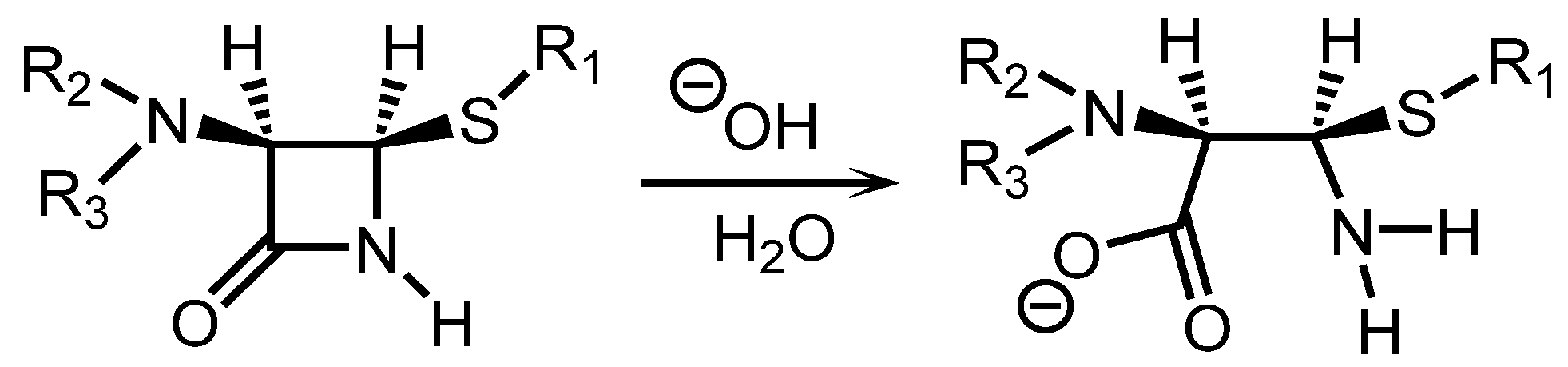
3.4. Increasing Affinity by Analyte Hydrolysis
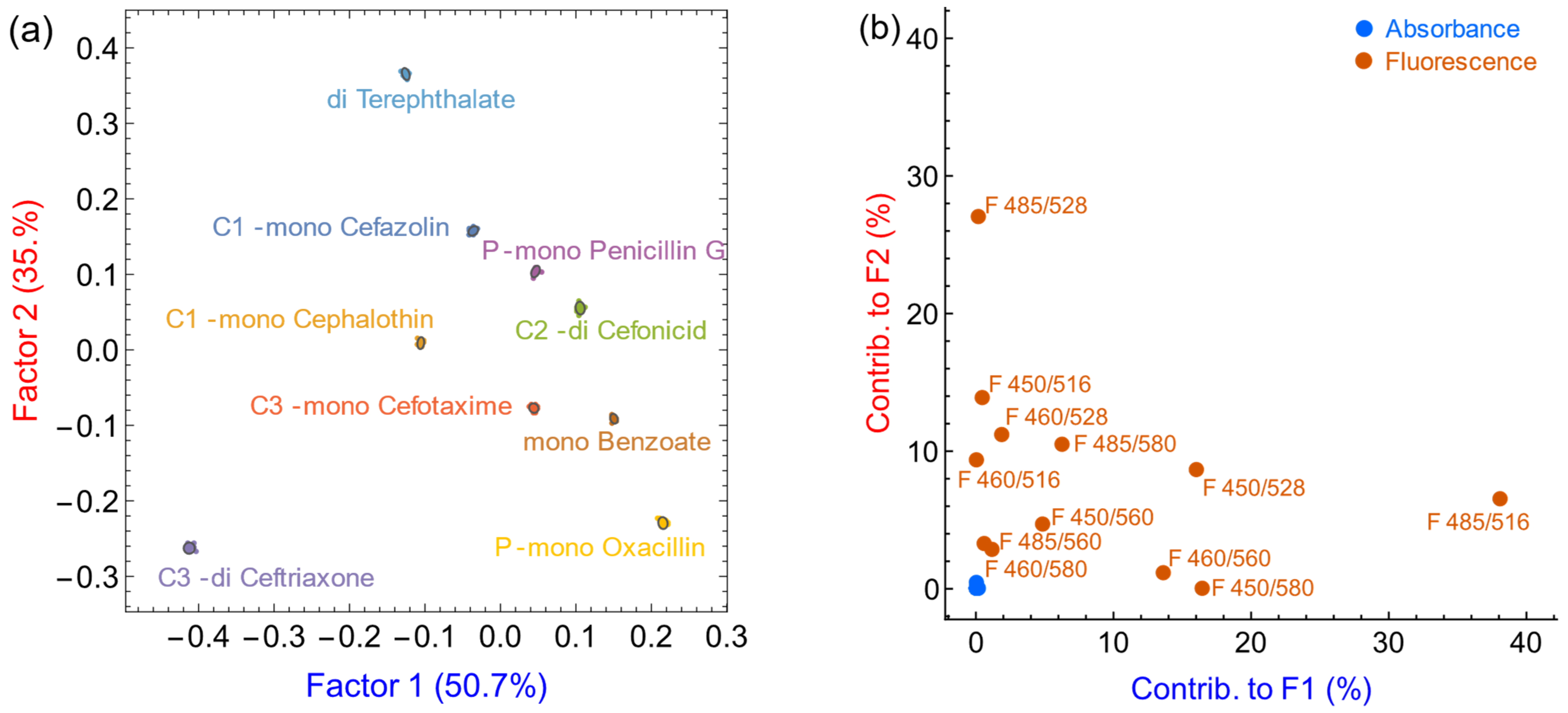
3.5. Antibiotic Discrimination on Partially Hydrolyzed Samples
3.6. Limit of Discrimination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munteanu, F.D.; Titoiu, A.M.; Marty, J.-L.; Vasilescu, A. Detection of antibiotics and evaluation of antibacterial activity with screen-printed electrodes. Sensors 2018, 18, 901. [Google Scholar] [CrossRef] [Green Version]
- Chafer-Pericas, C.; Maquieira, A.; Puchades, R. Fast screening methods to detect antibiotic residues in food samples. TrAC Trends Anal. Chem. 2010, 29, 1038–1049. [Google Scholar] [CrossRef]
- Rehman, M.S.U.; Rashid, N.; Ashfaq, M.; Saif, A.; Ahmad, N.; Han, J.-I. Global risk of pharmaceutical contamination from highly populated developing countries. Chemosphere 2015, 138, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elander, R.P. Industrial production of β-lactam antibiotics. Appl. Microbiol. Biotechnol. 2003, 61, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Delepierre, A.; Gayot, A.; Carpentier, A. Update on counterfeit antibiotics worldwide; public health risks. Med. Mal. Infect. 2012, 42, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Kelesidis, T.; Falagas, M.E. Substandard/counterfeit antimicrobial drugs. Clin. Microbiol. Rev. 2015, 28, 443–464. [Google Scholar] [CrossRef] [Green Version]
- Okerman, L.; Van Hoof, J.; Debeuckelaere, W. Evaluation of the European four-plate test as a tool for screening antibiotic residues in meat samples from retail outlets. J. AOAC Int. 1998, 81, 51–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oboegbulem, S.I.; Fidelis, A.P. Detection of antimicrobial residues in poultry meat and slaughter cattle in Nigeria. Meat Sci. 1996, 43, 71–74. [Google Scholar] [CrossRef]
- Collins-Thompson, D.L.; Wood, D.S.; Thomson, I.Q. Detection of antibiotic residues in consumer milk supplies in north America using the Charm test II procedure. J. Food Prot. 1988, 51, 632–633. [Google Scholar] [CrossRef]
- Rajkowski, K.T.; Peeler, J.T.; Messer, J.W. Detectability levels of four beta-lactam antibiotics in eight milk products using the AOAC Bacillus stearothermophilus Disc Assay. J. Food Prot. 1986, 49, 687–690. [Google Scholar] [CrossRef]
- Pikkemaat, M.G.; Dijk, S.O.-v.; Schouten, J.; Rapallini, M.; Kortenhoeven, L.; van Egmond, H.J. Nouws antibiotic test: Validation of a post-screening method for antibiotic residues in kidney. Food Control. 2009, 20, 771–777. [Google Scholar] [CrossRef]
- Pikkemaat, M.G.; Rapallini, M.L.B.A.; Dijk, S.O.-v.; Elferink, J.W.A. Comparison of three microbial screening methods for antibiotics using routine monitoring samples. Anal. Chim. Acta 2009, 637, 298–304. [Google Scholar] [CrossRef]
- Krisova, M.; Kozarova, I. Detection of residues of antimicrobial compounds in eggs by the rapid screening methods. Folia Vet. 2018, 62, 48–55. [Google Scholar] [CrossRef]
- Kozarova, I.; Juscakova, D.; simkova, J.; Milkovicova, M.; Kozar, M. Effective screening of antibiotic and coccidiostat residues in food of animal origin by reliable broadspectrum residue screening tests. Ital. J. Anim. Sci. 2020, 19, 487–501. [Google Scholar] [CrossRef]
- Okerman, L.; De Wasch, K.; Van Hoof, J. Detection of antibiotics in muscle tissue with microbiological inhibition tests: Effects of the matrix. Analyst 1998, 123, 2361–2365. [Google Scholar] [CrossRef] [PubMed]
- Pikkemaat, M.G.; Rapallini, M.L.B.A.; Zuidema, T.; Elferink, J.W.A.; Oostra-van Dijk, S.; Driessen-van Lankveld, W.D.M. Screening methods for the detection of antibiotic residues in slaughter animals: Comparison of the European Union Four-Plate Test, the Nouws Antibiotic Test and the Premi (R) Test (applied to muscle and kidney). Food Addit. Contam. Part A Chem. Anal. Control. 2011, 28, 26–34. [Google Scholar] [CrossRef]
- Jolly, A.M.; Bonizzoni, M. Intermolecular forces driving encapsulation of small molecules by PAMAM dendrimers in water. Macromolecules 2014, 47, 6281–6288. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. A New Class of Polymers: Starburst-Dendritic Macromolecules. Polym. J. 1985, 17, 117. [Google Scholar] [CrossRef] [Green Version]
- Tang, M.X.; Redemann, C.T.; Szoka, F.C. In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjugate Chem. 1996, 7, 703–714. [Google Scholar] [CrossRef]
- Niu, Y.; Sun, L.; Crooks, R.M. Determination of the intrinsic proton binding constants for poly(amidoamine) dendrimers via potentiometric pH titration. Macromolecules 2003, 36, 5725–5731. [Google Scholar] [CrossRef]
- Xu, Y.; Bonizzoni, M. Discrimination and quantitation of biologically relevant carboxylate anions using a [dye • PAMAM] complex. Sensors 2021, 21, 3637. [Google Scholar] [CrossRef]
- Westad, F.; Marini, F. Validation of chemometric models—A tutorial. Anal. Chim. Acta 2015, 893, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Bonizzoni, M.; Long, S.R.; Rainwater, C.; Anslyn, E.V. PAMAM dendrimer-induced aggregation of 5(6)-carboxyfluorescein. J. Org. Chem. 2012, 77, 1258–1266. [Google Scholar] [CrossRef] [PubMed]
- Jolly, A.M.; Bonizzoni, M. PAMAM dendrimers as supramolecular hosts through non-covalent interactions. Supramol. Chem. 2015, 27, 151–160. [Google Scholar] [CrossRef]
- Stewart, S.; Ivy, M.A.; Anslyn, E.V. The use of principal component analysis and discriminant analysis in differential sensing routines. Chem. Soc. Rev. 2014, 43, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, J.J.; Savoy, S.; Clevenger, M.B.; Ritchie, J.E.; McDoniel, B.; Yoo, S.-J.; Anslyn, E.V.; McDevitt, J.T.; Shear, J.B.; Neikirk, D. Solution-based analysis of multiple analytes by a sensor array: Toward the development of an “electronic tongue”. J. Am. Chem. Soc. 1998, 120, 6429–6430. [Google Scholar] [CrossRef]
- Collins, B.E.; Anslyn, E.V. Pattern-based peptide recognition. Chem. Eur. J. 2007, 13, 4700–4708. [Google Scholar] [CrossRef]
- Roggo, Y.; Duponchel, L.; Huvenne, J.P. Comparison of supervised pattern recognition methods with McNemar’s statistical test. Application to qualitative analysis of sugar beet by near-infrared spectroscopy. Anal. Chim. Acta 2003, 477, 187–200. [Google Scholar] [CrossRef]
- Bocklitz, T.; Richard, G. Brereton: Chemometrics: Data driven extraction for science, 2nd ed. Anal. Bioanal. Chem. 2019, 411, 2995–2996. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Jia, X.-D.; Gao, R.-X.; Chen, M.-L.; Yang, T.; Wang, J.-H. Discrimination of pathogenic bacteria with boronic acid modified protonated g-C3N4 nanosheets at various pHs. Sens. Actuators B 2021, 340, 129951. [Google Scholar] [CrossRef]
- Zhou, W.; Hou, J.; Li, Y.; Zhou, H.; Huang, H.; Zhang, L.; Nawaz, M.A.H.; Yu, C. Protein discrimination based on DNA induced perylene probe self-assembly. Talanta 2021, 224, 121897. [Google Scholar] [CrossRef]
- Geana, E.-I.; Ciucure, C.T.; Apetrei, C. Electrochemical sensors coupled with multivariate statistical analysis as screening tools for wine authentication issues: A review. Chemosensors 2020, 8, 59. [Google Scholar] [CrossRef]
- Liang, X.; Bonizzoni, M. Boronic acid-modified poly(amidoamine) dendrimers as sugar-sensing materials in water. J. Mater. Chem. B 2016, 4, 3094–3103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.; Trentle, M.; Kozlovskaya, V.; Kharlampieva, E.; Bonizzoni, M. Carbohydrate sensing using water-soluble poly(methacrylic acid)-co-3-(acrylamido)phenylboronic acid copolymer. ACS Appl. Polym. Mater. 2019, 1, 1341–1349. [Google Scholar] [CrossRef]
- Ihde, M.H.; Pridmore, C.F.; Bonizzoni, M. Pattern-based recognition systems: Overcoming the problem of mixtures. Anal. Chem. 2020, 92, 16213–16220. [Google Scholar] [CrossRef]
- Mallet, A.M.; Davis, A.B.; Davis, D.R.; Panella, J.; Wallace, K.J.; Bonizzoni, M. A cross reactive sensor array to probe divalent metal ions. Chem. Commun. 2015, 51, 16948–16951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carneiro, S.V.; Holanda, M.H.B.; Cunha, H.O.; Oliveira, J.J.P.; Pontes, S.M.A.; Cruz, A.A.C.; Fechine, L.M.U.D.; Moura, T.A.; Paschoal, A.R.; Zambelli, R.A.; et al. Highly sensitive sensing of food additives based on fluorescent carbon quantum dots. J. Photochem. Photobiol. A 2021, 411, 113198. [Google Scholar] [CrossRef]
- Mitchell, L.; Shen, C.; Timmins, H.C.; Park, S.B.; New, E.J. A versatile fluorescent sensor array for platinum anticancer drug detection in biological fluids. ACS Sens. 2021, 6, 1261–1269. [Google Scholar] [CrossRef] [PubMed]



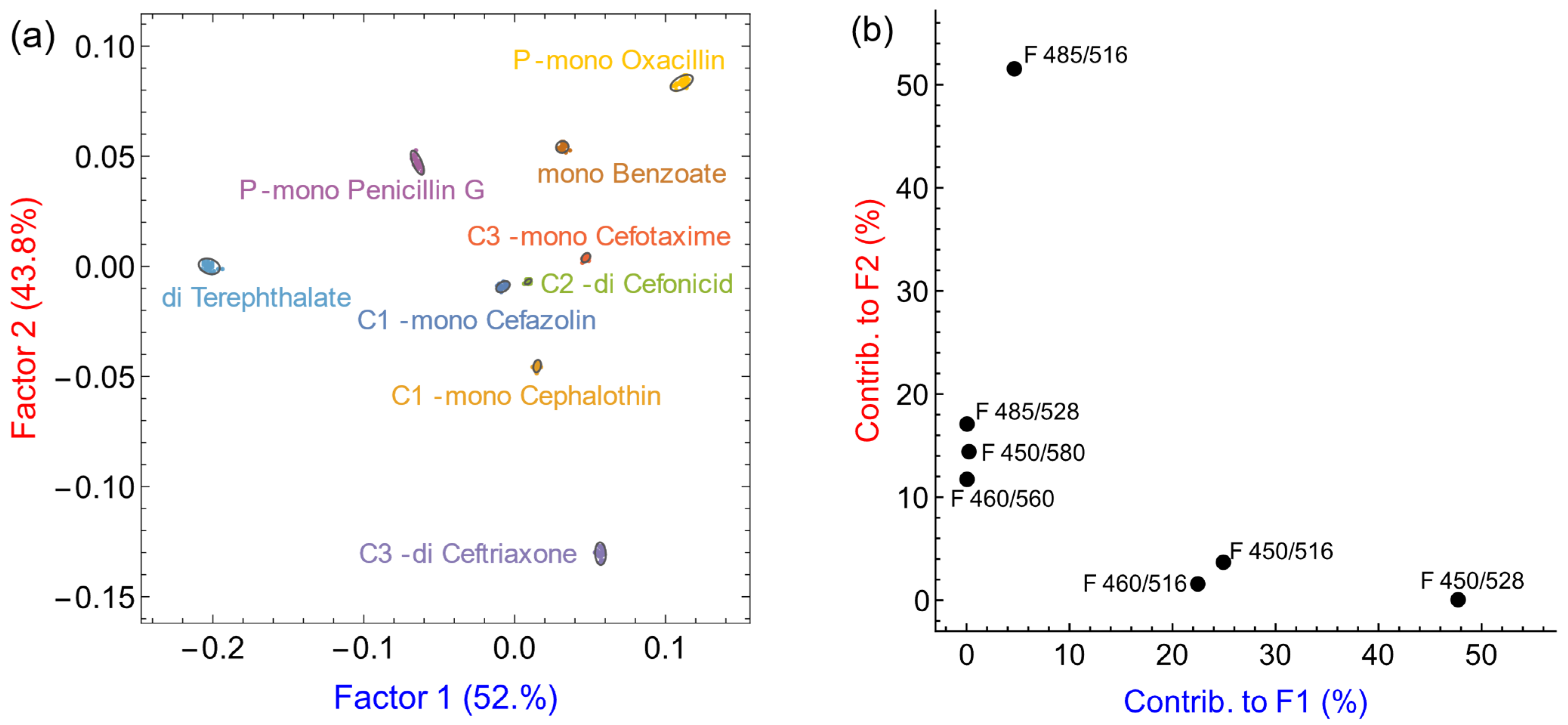
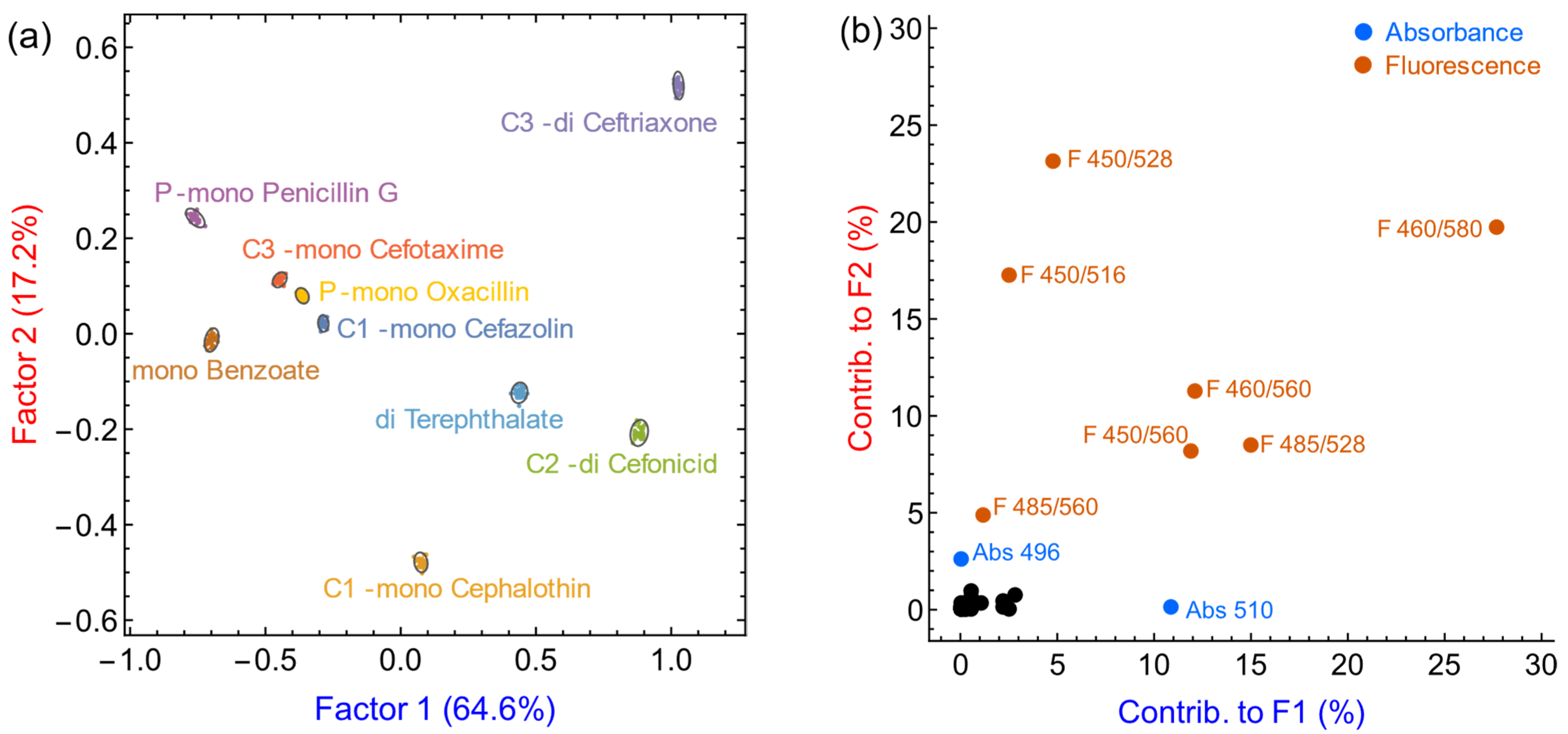
| Antibiotic | Family | Nominal Charge | Relative Affinity K |
|---|---|---|---|
| Cefazolin | cephalosporin | monoanion | 115 ± 7 |
| Cefotaxime | cephalosporin | monoanion | 86 ± 8 |
| Cephalothin | cephalosporin | monoanion | 5068 ± 262 1 |
| Penicillin G | penicillin | monoanion | 81 ± 9 |
| Oxacillin | penicillin | monoanion | 58 ± 5 |
| Cefonicid | cephalosporin | dianion | 1073 ± 118 |
| Ceftriaxone | cephalosporin | dianion | 1251 ± 97 |
| Terephthalate | reference | dianion | 590 ± 102 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Bonizzoni, M. Beta-Lactam Antibiotic Discrimination Using a Macromolecular Sensor in Water at Neutral pH. Sensors 2021, 21, 6384. https://doi.org/10.3390/s21196384
Xu Y, Bonizzoni M. Beta-Lactam Antibiotic Discrimination Using a Macromolecular Sensor in Water at Neutral pH. Sensors. 2021; 21(19):6384. https://doi.org/10.3390/s21196384
Chicago/Turabian StyleXu, Yifei, and Marco Bonizzoni. 2021. "Beta-Lactam Antibiotic Discrimination Using a Macromolecular Sensor in Water at Neutral pH" Sensors 21, no. 19: 6384. https://doi.org/10.3390/s21196384