Recognition of EEG Signals from Imagined Vowels Using Deep Learning Methods
Abstract
:1. Introduction
2. Related Work
3. Materials and Methods
3.1. Data Description
3.1.1. Reference Database (BD1)
3.1.2. New Database (BD2)
3.2. Deep Learning Methods with Convolutional Neural Networks (CNN)
3.2.1. CNNeeg1-1 Architecture
Preprocessing
- For each multidimensional input frame and each shift operation (t), decompose the covariance matrix as where is the eigenvector matrix, and is the eigenvalue matrix. In this case the largest eigenvalue will correspond to the eigenvector .
- Take the first principal component and build a vector pointing in the opposite direction to .
- Using the Hammerseley sequence on a uniformly sampled sphere, build a set of K direction vectors .
- Calculate the Euclidean distances from each of the uniform direction vectors to .
- Relocate half of the projection vectors , the closest to using =, where is used to control the density of the relocated vectors.
- The other half of the uniform projection vectors, , the closest to , are relocated using =, where is used to control the density of the relocated vectors.
- Project the multidimensional signal along the direction vectors found in steps 5 and 6.
- Find the instant of time corresponding to the maximum of the projected data sets, where is the angle of the dimensional sphere and is the index of the direction vectors.
- Interpolate to calculate the envelope curves .
- Estimate the mean of the envelope curves for the set of direction vectors :
- Calculate the residue .
- Repeat these steps until the residue meets the conditions of an IMF for multivariate signals.
CNNeeg1-1: A New Deep Learning Architecture with CNN
4. Results
4.1. Analysis of Intra-Subject Training Results for the Shallow CNN, EEGNet, and CNNeeg1-1 Algorithms Using Databases BD1 and BD2
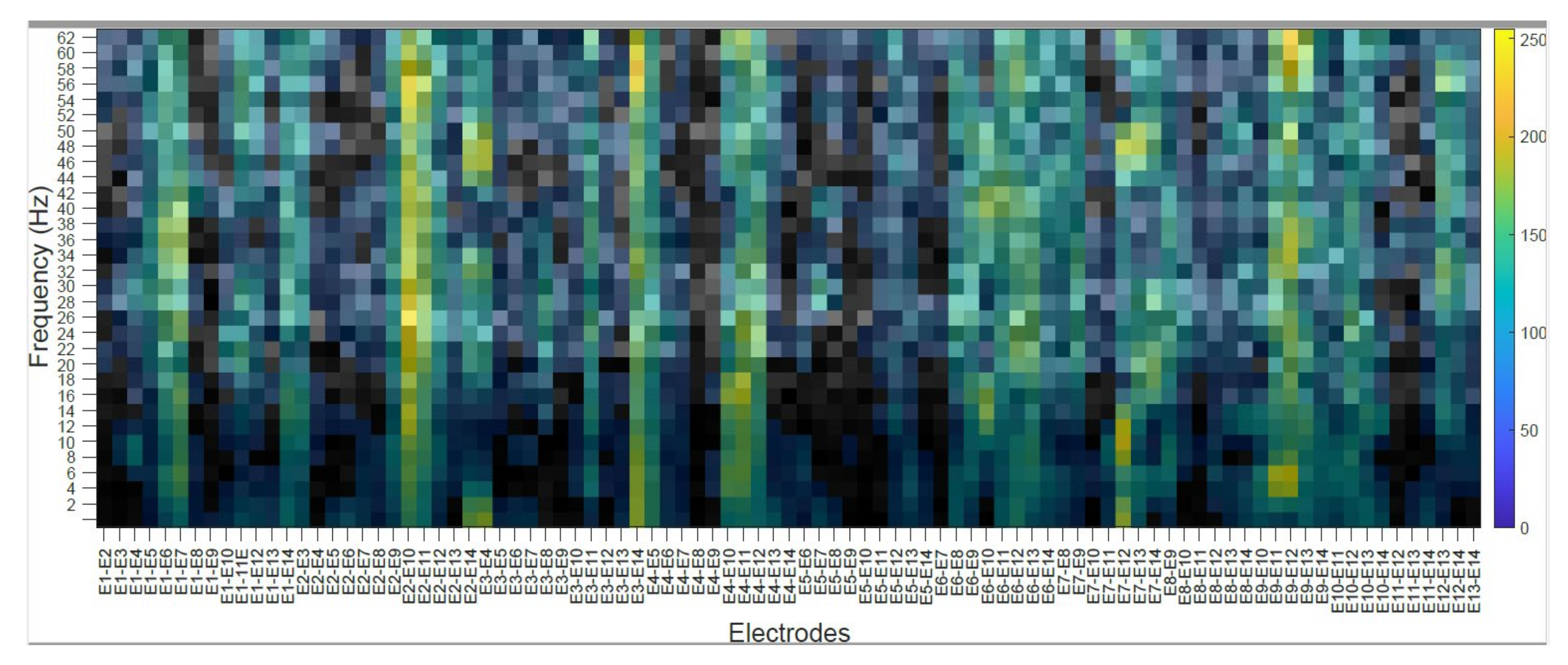
4.2. Subject’s Internal Visualization BD2 Database CNNeeg1-1
4.3. Analysis of the Inter-Subject Training Results for the Shallow CNN, EEGNet, and CNNeeg1-1 Algorithms Using BD1 and BD2 Databases
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BCI | Brain-computer interfaces |
| EEG | Electroencephalogram |
| IS | Imagined speech |
| ML | Machine learning |
| DL | Deep Learning |
| CNN | Convolutional neural networks |
| BD1 | Reference database |
| BD2 | New database |
| APIT-MEMD | Adaptive-Projection Intrinsically Transformed MEMD |
| CAM | Class Activation Mapping |
| ERP | Event Related Potential |
| MRCP | Movement Related Cortical Potential |
| SMR | Sensorimotor rhythms |
| ERS/ERD | Event-related synchronization/desynchronization |
| MI | Motor imagery |
| SVM | Support Vector Machine |
| CSPs | Common special patterns |
| RF | Random Forest |
| LDA | Linear Discriminant Analysis |
| AC | Adaptive collection |
| DWT | Discrete Wavelet Transform |
| DBN | Deep Belief Networks |
Appendix A. (Shallow CNN Architecture)

Appendix B. (EEGNet Architecture)

References
- Sarmiento, L.C. Interfaces Cerebro-Computador para el Reconocimiento Automático del Habla Silenciosa; Universidad Pedagógica Nacional: Bogotá, Colombia, 2019; pp. 37–39. [Google Scholar]
- Han, C.H.; Hwang, H.J.; Im, C.H. Classification of visual stimuli with different spatial patterns for single-frequency, multi-class ssvep bci. Electron. Lett. 2013, 49, 1374–1376. [Google Scholar] [CrossRef]
- Ha, K.W.; Jeong, J.W. Motor imagery EEG classification using capsule networks. Sensors 2019, 19, 2854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Zheng, H.; Wang, J.; Li, D.; Fang, X. Recognition of EEG signal motor imagery intention based on deep multi-view feature learning. Sensors 2020, 20, 3496. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Laine, T.H.; Sohn, K.A. An exploration of machine learning methods for robust boredom classification using EEG and GSR data. Sensors 2019, 19, 4561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerous, B.; Skola, F.; Liarokapis, F. EEG-based BCI and video games: A progress report. Virtual Real. 2018, 22, 119–135. [Google Scholar] [CrossRef]
- Li, X.; Chen, X.; Yan, Y.; Wei, W.; Wang, Z.J. Classification of EEG signals using a multiple kernel learning support vector machine. Sensors 2014, 14, 12784–12802. [Google Scholar] [CrossRef] [Green Version]
- Tayeb, Z.; Fedjaev, J.; Ghaboosi, N.; Richter, C.; Everding, L.; Qu, X.; Conradt, J. Validating deep neural networks for online decoding of motor imagery movements from EEG signals. Sensors 2019, 19, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Xu, G.; Han, Z.; Ma, K.; Zheng, X.; Chen, L.; Zhang, S. Data augmentation for motor imagery signal classification based on a hybrid neural network. Sensors 2020, 20, 4485. [Google Scholar] [CrossRef]
- Brigham, K.; Kumar, B.V. Imagined speech classification with EEG signals for silent communication: A preliminary investigation into synthetic telepathy. In Proceedings of the 2010 4th International Conference on Bioinformatics and Biomedical Engineering, Chengdu, China, 23 July 2010. [Google Scholar]
- Ikeda, S.; Shibata, T.; Nakano, N.; Okada, R.; Tsuyuguchi, N.; Ikeda, K.; Kato, A. Neural decoding of single vowels during covert articulation using electrocorticography. Front. Hum. Neurosci. 2014, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Morooka, T.; Ishizuka, K.; Kobayashi, N. Electroencephalographic analysis of auditory imagination to realize silent speech BCI. In Proceedings of the 2018 IEEE 7th Global Conference on Consumer Electronics (GCCE 2018), Nara, Japan, 9–12 October 2018; pp. 683–686. [Google Scholar]
- Cooney, C.; Folli, R.; Coyle, D. Neurolinguistics Research Advancing Development of a Direct-Speech Brain-Computer Interface. IScience 2018, 8, 103–125. [Google Scholar] [CrossRef] [Green Version]
- Kamalakkannan, R.; Rajkumar, R.; Madan, R.M.; Shenbaga, D.S. Imagined Speech Classification using EEG. Adv. Biomed. Sci. Eng. 2014, 1, 20–32. [Google Scholar]
- Matsumoto, M.; Hori, J. Classification of silent speech using support vector machine and relevance vector machine. Appl. Soft Comput. 2014, 20, 95–102. [Google Scholar] [CrossRef]
- Coretto, G.A.; Gareis, I.E.; Rufiner, H.L. Open access database of EEG signals recorded during imagined speech. In Proceedings of the 12th International Symposium on Medical Information, Processing and Analysis International Society for Optics and Photonics, Tandil, Argentina, 26 January 2017. [Google Scholar]
- Cooney, C.; Korik, A.; Folli, R.; Coyle, D. Evaluation of hyperparameter optimization in machine and deep learning methods for decoding imagined speech EEG. Sensors 2020, 20, 4629. [Google Scholar] [CrossRef]
- Lee, T.J.; Sim, K.B. Vowel classification of imagined speech in an electroencephalogram using the deep belief network. J. Inst. Control. Robot. Syst. 2015, 21, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Min, B.; Kim, J.; Park, H.J.; Lee, B. Vowel imagery decoding toward silent speech BCI using extreme learning machine with electroencephalogram. BioMed Res. Int. 2016, 2016, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Hickok, G.; Poeppel, D. The cortical organization of speech processing. Nat. Rev. Neurosci. 2007, 8, 393–402. [Google Scholar] [CrossRef] [PubMed]
- He, B. Focused ultrasound help realize high spatiotemporal brain imaging-A concept on acoustic-electrophysiological neuroimaging. IEEE Trans. Biomed. Eng. 2016, 63, 2654–2656. [Google Scholar]
- Ukil, A. Denoising and frequency analysis of noninvasive magnetoencephalography sensor signals for functional brain mapping. IEEE Sens. J. 2010, 12, 447–455. [Google Scholar] [CrossRef]
- Jeong, S.; Li, X.; Yang, J.; Li, Q.; Tarokh, V. Sparse representation-based denoising for high-resolution brain activation and functional connectivity modeling: A task fMRI study. IEEE Access 2020, 8, 36728–36740. [Google Scholar] [CrossRef]
- Jaber, H.A.; Aljobouri, H.K.; Çankaya, İ.; Koçak, O.M.; Algin, O. Preparing fMRI data for postprocessing: Conversion modalities, preprocessing pipeline, and parametric and nonparametric approaches. IEEE Access 2019, 7, 122864–122877. [Google Scholar] [CrossRef]
- Mullen, T.R.; Kothe, C.A.; Chi, Y.M.; Ojeda, A.; Kerth, T.; Makeig, S.; Cauwenberghs, G. Real-time neuroimaging and cognitive monitoring using wearable dry EEG. IEEE Trans. Biomed. Eng. 2015, 62, 2553–2567. [Google Scholar] [CrossRef] [Green Version]
- Ten Caat, M.; Maurits, N.M.; Roerdink, J.B. Data-driven visualization and group analysis of multichannel EEG coherence with functional units. IEEE Trans. Vis. Comput. Graph. 2008, 14, 756–771. [Google Scholar] [CrossRef]
- Sanei, S.; Chambers, J.A. EEG Signal Processing; John Wiley & Sons: Chichester, UK, 2013; pp. 28–35. [Google Scholar]
- Rashid, M.; Sulaiman, N.; Majeed, A.P.; Musa, R.M.; Nasir, A.F.; Bari, B.S.; Khatun, S. Current status, challenges, and possible solutions of EEG-based brain-computer interface: A comprehensive review. Front. Neurorobot. 2020, 14, 25. [Google Scholar] [CrossRef]
- Graimann, B.; Allison, B.Z.; Pfurtscheller, G. Brain–Computer Interfaces: Revolutionizing Human–Computer Interaction; Springer: Berlin/Heidelberg, Germany, 2010; pp. 137–154. [Google Scholar]
- Khan, S.M.; Khan, A.A.; Farooq, O. Selection of features and classifiers for EMG-EEG-based upper limb assistive devices—A review. IEEE Rev. Biomed. Eng. 2019, 13, 248–260. [Google Scholar] [CrossRef]
- Lampropoulos, A.S.; Tsihrintzis, G.A. Machine Learning Paradigms. Applications in Recommender Systems; Springer: Cham, Switzerland, 2015; Volume 92, pp. 101–115. [Google Scholar]
- Hosseini, M.P.; Hosseini, A.; Ahi, K. A review on machine learning for EEG signal processing in bioengineering. IEEE Rev. Biomed. Eng. 2020, 14, 204–2018. [Google Scholar] [CrossRef]
- Alpaydin, E. Introduction to Machine Learning, 4th ed.; MIT Press: Cambridge, MA, USA, 2020; pp. 313–318. [Google Scholar]
- Müller, K.R.; Tangermann, M.; Dornhege, G.; Krauledat, M.; Curio, G.; Blankertz, B. Machine learning for real-time single-trial EEG-analysis: From brain–computer interfacing to mental state monitoring. J. Neurosci. Methods 2008, 167, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Kamath, U.; Liu, J.; Whitaker, J. Deep Learning for NLP and Speech Recognition; Springer: Berlin/Heidelberg, Germany, 2019; pp. 263–289. [Google Scholar]
- Roy, Y.; Banville, H.; Albuquerque, I.; Gramfort, A.; Falk, T.H.; Faubert, J. Deep learning-based electroencephalography analysis: A systematic review. J. Neural Eng. 2019, 16, 051001. [Google Scholar] [CrossRef]
- Campesato, O. Artificial Intelligence, Machine Learning, and Deep Learning; Mercury Learning and Information LLC: Boston, MA, USA, 2020; pp. 55–73. [Google Scholar]
- Gulli, A.; Kapoor, A.; Pal, S. Deep Learning with TensorFlow 2 and Keras: Regression, ConvNets, GANs, RNNs, NLP, and More with TensorFlow 2 and the Keras API, 2nd ed.; Packt Publishing Ltd.: Birmingham, UK, 2019; pp. 109–191, 279–340. [Google Scholar]
- Samek, W.; Montavon, G.; Lapuschkin, S.; Anders, C.J.; Müller, K.R. Explaining deep neural networks and beyond: A review of methods and applications. Proc. IEEE 2021, 109, 247–278. [Google Scholar] [CrossRef]
- Chengaiyan, S.; Retnapandian, A.S.; Anandan, K. Identification of vowels in consonant–vowel–consonant words from speech imagery based EEG signals. Cogn. Neurodyn. 2020, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tamm, M.O.; Muhammad, Y.; Muhammad, N. Classification of vowels from imagined speech with convolutional neural networks. Computers 2020, 9, 46. [Google Scholar] [CrossRef]
- Cooney, C.; Raffaella, F.; Coyle, D. Optimizing input layers improves CNN generalization and transfer learning for imagined speech decoding from EEG. In Proceedings of the 2019 IEEE International Conference on Systems, Man and Cybernetics (SMC 2019), Bari, Italy, 6–9 October 2019. [Google Scholar]
- Reader, A.J.; Corda, G.; Mehranian, A.; da Costa-Luis, C.; Ellis, S.; Schnabel, J.A. Deep learning for PET image reconstruction. IEEE Trans. Radiat. Plasma Med. Sci. 2020, 5, 1–25. [Google Scholar] [CrossRef]
- DiSpirito, A., III; Li, D.; Vu, T.; Chen, M.; Zhang, D.; Luo, J.; Yao, J. Reconstructing undersampled photoacoustic microscopy images using deep learning. IEEE Trans. Med. Imaging 2020, 40, 562–570. [Google Scholar] [CrossRef]
- Rostami, M.; Kolouri, S.; Murez, Z.; Owekcho, Y.; Eaton, E.; Kim, K. Zero-shot image classification using coupled dictionary embedding. arXiv 2019, arXiv:1906.10509. [Google Scholar]
- Barr, P.J.; Ryan, J.; Jacobson, N.C. Precision assessment of COVID-19 phenotypes using large-scale clinic visit audio recordings: Harnessing the power of patient voice. J. Med. Internet Res. 2021, 23, 20545. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Seong, J.J.; Ozlu, B.; Shim, B.S.; Marakhimov, A.; Lee, S. Biosignal sensors and deep learning-based speech recognition: A review. Sensors 2021, 21, 1399. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Li, K.; Ge, F.; Huang, Z.; Yang, M.; Siniscalchi, S.M.; Lee, C.H. An end-to-end deep learning approach to simultaneous speech dereverberation and acoustic modeling for robust speech recognition. IEEE J. Sel. Top. Signal Process. 2017, 11, 1289–1300. [Google Scholar] [CrossRef]
- Sereshkeh, A.R.; Trott, R.; Bricout, A.; Chau, T. EEG classification of covert speech using regularized neural networks. IEEE/ACM Trans. Audio Speech Lang. Process. 2017, 25, 2292–2300. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, M.; Lee, S.W. Neural decoding of imagined speech and visual imagery as intuitive paradigms for BCI communication. IEEE Trans. Neural Syst. Rehabil. Eng. 2020, 28, 2647–2659. [Google Scholar] [CrossRef]
- Sheykhivand, S.; Mousavi, Z.; Rezaii, T.Y.; Farzamnia, A. Recognizing emotions evoked by music using CNN-LSTM networks on EEG signals. IEEE Access 2020, 8, 139332–139345. [Google Scholar] [CrossRef]
- Hagad, J.L.; Kimura, T.; Fukui, K.I.; Numao, M. Learning subject-generalized topographical EEG embeddings using deep variational autoencoders and domain-adversarial regularization. Sensors 2021, 21, 1792. [Google Scholar] [CrossRef]
- Hemakom, A.; Goverdovsky, V.; Looney, D.; Mandic, D.P. Adaptive-projection intrinsically transformed multivariate empirical mode decomposition in cooperative brain–computer interface applications. Philos. Trans. R. Soc. A 2016, 374, 20150199. [Google Scholar] [CrossRef]
- Tan, P.N.; Steinbach, M.; Kumar, V. Introduction to Data Mining, 1st ed.; Pearson Education, Inc.: New Delhi, India, 2016; pp. 302–308. [Google Scholar]
- Wang, H.; Du, M.; Yang, F.; Zhang, Z. Score-cam: Improved visual explanations via score-weighted class activation mapping. arXiv 2019, arXiv:1910.01279. [Google Scholar]
- Singh, A.; Hussain, A.A.; Lal, S.; Guesgen, H.W. Comprehensive review on critical issues and possible solutions of motor imagery based electroencephalography brain-computer interface. Sensors 2021, 21, 2173. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Jiang, L.; Dong, G.; Gao, X.; Wang, Y. An open dataset for wearable SSVEP-based brain-computer interfaces. Sensors 2021, 21, 1256. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Nguyen, T.H.; Chung, W.Y. A bipolar-channel hybrid brain-computer interface system for home automation control utilizing steady-state visually evoked potential and eye-blink signals. Sensors 2020, 20, 5474. [Google Scholar] [CrossRef]
- Choi, J.; Kim, K.T.; Jeong, J.H.; Kim, L.; Lee, S.J.; Kim, H. Developing a motor imagery-based real-time asynchronous hybrid BCI controller for a lower-limb exoskeleton. Sensors 2020, 20, 7309. [Google Scholar] [CrossRef]
- Chailloux, J.D.; Mendoza, O.; Antelis, J.M. Single-option P300-BCI performance is affected by visual stimulation conditions. Sensors 2020, 20, 7198. [Google Scholar] [CrossRef]
- Li, M.; Li, F.; Pan, J.; Zhang, D.; Zhao, S.; Li, J.; Wang, F. The MindGomoku: An online P300 BCI game based on bayesian deep learning. Sensors 2021, 21, 1613. [Google Scholar] [CrossRef]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A compact convolutional neural network for EEG-based brain–computer interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef] [Green Version]














| Classifiers | Accuracy | Subjects | Electrodes |
|---|---|---|---|
| Support Vector Machine with Gaussian kernel (SVM-G) [15] | 77% | 5 | 19 |
| Relevance Vector Machine with Gaussian kernel (RVM-G) [15] | 79% | 5 | 19 |
| Linear Relevance Vector Machine (RVM-L) [15] | 50% | 5 | 19 |
| Bipolar Neural Network [14] | 44% | 13 | 19 |
| Support Vector Machine (SVM) [16] | 21.94% | 15 | 6 |
| Random Forest (RF) [16] | 22.72% | 15 | 6 |
| Extreme Learning Machine (ELM) [19] | 57–82% | 5 | 64 |
| Extreme Learning Machine with Linear Function (ELM-L) [19] | 60–85% | 5 | 64 |
| Extreme Learning Machine with Radial Basis Function (ELM-R) [19] | 62–85% | 5 | 64 |
| Support Vector Machine with Radial Basis Function Kernel (SVM-R) [19] | 50–55% | 5 | 64 |
| Linear Discriminant Analysis (LDA) [19] | 55–80% | 5 | 64 |
| SVM [17] | 22.23% | 15 | 6 |
| Random Forest [17] | 23.08% | 15 | 6 |
| rLDA [17] | 25.82% | 15 | 6 |
| DL Architecture | Accuracy | Subjects | Electrodes |
|---|---|---|---|
| Deep Belief Networks (DBN) [40] | 80% | 6 | 19 |
| Deep Belief Networks (DBN) [18] | 87.96% | 3 | 32 |
| Recurrent Neural Networks (RNN) [40] | 70% | 6 | 19 |
| Convolutional Neural Networks (CNN) [41] | 32.75% | 15 | 6 |
| Convolutional Neural Networks (CNN) [42] | 35.68% | 15 | 6 |
| Shallow CNN [17] | 29.62% | 15 | 6 |
| Deep CNN [17] | 29.06% | 15 | 6 |
| EEGNet [17] | 30.08% | 15 | 6 |
| Name | Type | Activations | Learnables | Total Learnables | |
|---|---|---|---|---|---|
| 1 | Input 32 × 91 × 1 images with ‘zerocenter’ normalization | Image input | 32 × 91 × 1 | - | 0 |
| 2 | dropout 25% dropout | Dropout | 32 × 91 × 1 | - | 0 |
| 3 | conv_1 50 5 × 5 × 1 convolutions with stride 1 × 1 and p… | Convolution | 28 × 87 × 50 | Weights 5 × 5 × 1 × 50 Bias 1 × 1 × 50 | 1300 |
| 4 | BN_1 Batch normalization with 50 channels | Batch Normalization | 28 × 87 × 50 | Offset 1 × 1 × 50 Scale 1 × 1 × 50 | 100 |
| 5 | relu_1 ReLU | ReLU | 28 × 87 × 50 | - | 0 |
| 6 | pool_1 2 × 2 max pooling with stride 2 × 2 and padding… | Max Pooling | 14 × 43 × 50 | - | 0 |
| 7 | conv_2 60 11 × 11 × 50 convolutions with stride 1 × 1 an | Convolution | 4 × 33 × 60 | Weights 11 × 11 × 50 × 50 Bias 1 × 1 × 60 | 363,060 |
| 8 | BN_2 Batch normalization with 60 channels | Batch Normalization | 4 × 33 × 60 | Offset 1 × 1 × 60 Scale 1 × 1 × 60 | 120 |
| 9 | relu_2 ReLU | ReLU | 4 × 33 × 60 | - | 0 |
| 10 | pool_2 2 × 2 max pooling with stride 2 × 2 and padding… | Max Pooling | 2 × 16 × 60 | - | 0 |
| 11 | BN_3 Batch normalization with 60 channels | Batch Normalization | 2 × 16 × 60 | Offset 1 × 1 × 60 Scale 1 × 1 × 60 | 120 |
| 12 | fc1 60 fully connected layer | Fully Connected | 1 × 1 × 60 | Weights 60×1920 Bias 60×1 | 115,260 |
| 13 | fc2 2 fully connected layer | Fully Connected | 1 × 1 × 2 | Weights 2 × 60 Bias 2 × 1 | 122 |
| 14 | softmax softmax | softmax | 1 × 1 × 2 | - | 0 |
| 15 | classOutput crossentropyex | Classification Output | - | - | 0 |
| Shallow CNN (BD1) | EEGNet (BD1) | CNNeeg1-1 (BD1) | ||||
|---|---|---|---|---|---|---|
| Model Training | Intra | Inter | Intra | Inter | Intra | Inter |
| Mean | 0.3171 | 0.2587 | 0.3506 | 0.3531 | 0.6562 | 0.5008 |
| SD | 0.0114 | 0.0157 | 0.0133 | 0.2774 | 0.0123 | 0.0133 |
| Shallow CNN (BD2) | EEGNet (BD2) | CNNeeg1-1 (BD2) | ||||
|---|---|---|---|---|---|---|
| Model Training | Intra | Inter | Intra | Inter | Intra | Inter |
| Mean | 0.5371 | 0.2475 | 0.7068 | 0.4578 | 0.8566 | 0.6276 |
| SD | 0.0606 | 0.0245 | 0.0396 | 0.0433 | 0.0446 | 0.0644 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarmiento, L.C.; Villamizar, S.; López, O.; Collazos, A.C.; Sarmiento, J.; Rodríguez, J.B. Recognition of EEG Signals from Imagined Vowels Using Deep Learning Methods. Sensors 2021, 21, 6503. https://doi.org/10.3390/s21196503
Sarmiento LC, Villamizar S, López O, Collazos AC, Sarmiento J, Rodríguez JB. Recognition of EEG Signals from Imagined Vowels Using Deep Learning Methods. Sensors. 2021; 21(19):6503. https://doi.org/10.3390/s21196503
Chicago/Turabian StyleSarmiento, Luis Carlos, Sergio Villamizar, Omar López, Ana Claros Collazos, Jhon Sarmiento, and Jan Bacca Rodríguez. 2021. "Recognition of EEG Signals from Imagined Vowels Using Deep Learning Methods" Sensors 21, no. 19: 6503. https://doi.org/10.3390/s21196503
APA StyleSarmiento, L. C., Villamizar, S., López, O., Collazos, A. C., Sarmiento, J., & Rodríguez, J. B. (2021). Recognition of EEG Signals from Imagined Vowels Using Deep Learning Methods. Sensors, 21(19), 6503. https://doi.org/10.3390/s21196503






