Automatic Cephalometric Landmark Identification System Based on the Multi-Stage Convolutional Neural Networks with CBCT Combination Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Image Acquisition from CBCT
2.2.1. CBCT Protocol
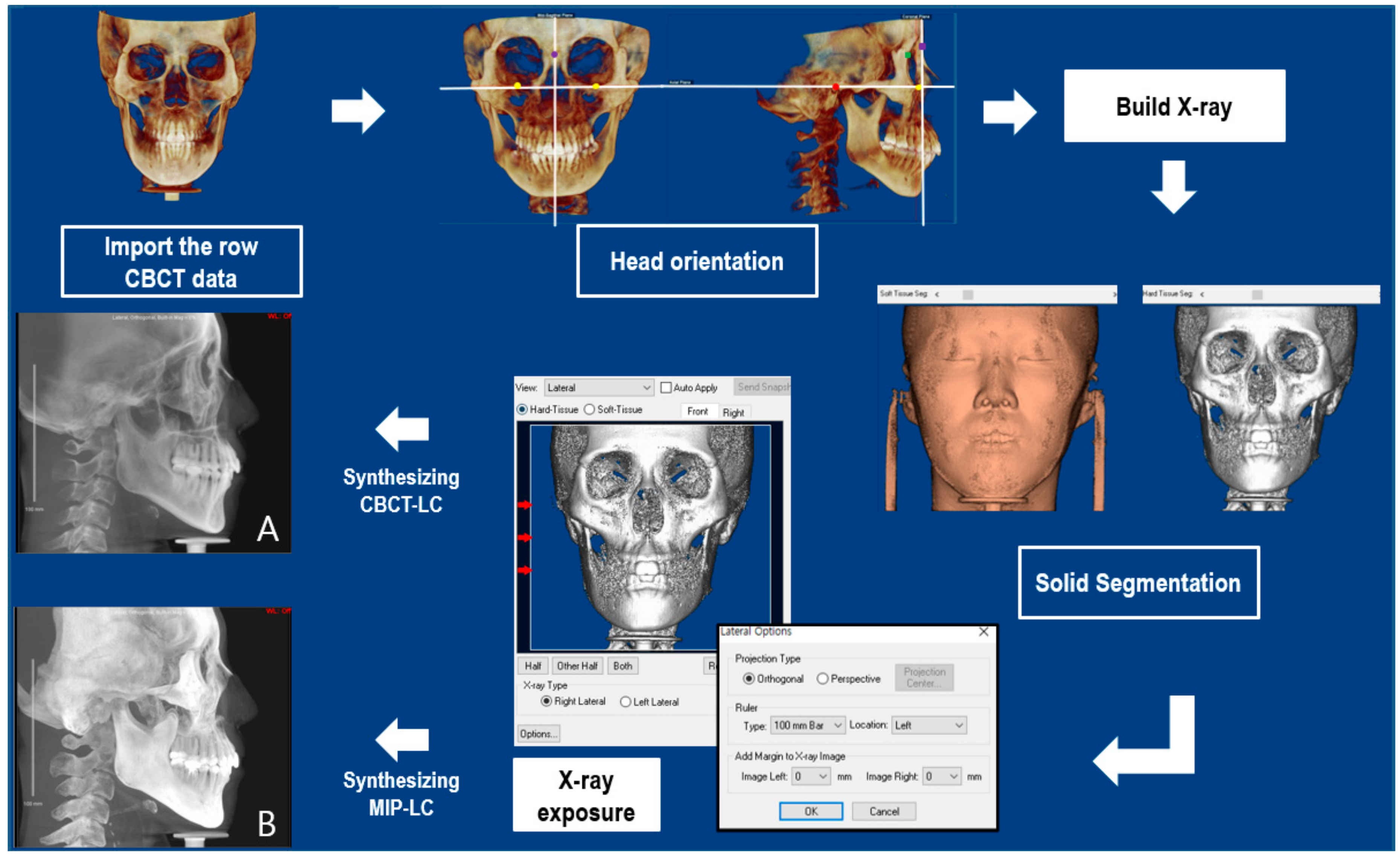
2.2.2. Reorientation
2.2.3. Synthesizing the Cephalogram
2.3. Reproducibility of Intra-Examiner
2.4. Multi-Stage CNNs Architecture
2.5. System Evaluation
2.6. AI Prediction on Different Lateral Cephalograms
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smektala, T.; Jedrzejewski, M.; Szyndel, J.; Sporniak-Tutak, K.; Olszewski, R. Experimental and clinical assessment of three-dimensional cephalometry: A systematic review. J. Craniomaxillofac. Surg. 2014, 42, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Gribel, B.F.; Gribel, M.N.; Frazao, D.C.; McNamara, J.A., Jr.; Manzi, F.R. Accuracy and reliability of craniometric measurements on lateral cephalometry and 3D measurements on CBCT scans. Angle Orthod. 2011, 81, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.H.; Kim, M.J.; Shin, H.J.; Cho, H.J.; Baek, S.H. Three-dimensional surgical accuracy between virtually planned and actual surgical movements of the maxilla in two-jaw orthognathic surgery. Korean J. Orthod. 2020, 50, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Kapila, S.D.; Nervina, J.M. CBCT in orthodontics: Assessment of treatment outcomes and indications for its use. Dentomaxillofac. Radiol. 2015, 44, 20140282. [Google Scholar] [CrossRef]
- Na, E.R.; Aljawad, H.; Lee, K.M. A comparative study of the reproducibility of landmark identification on posteroanterior and anteroposterior cephalograms generated from cone-beam computed tomography scans. Korean J. Orthod. 2019, 49, 41–48. [Google Scholar] [CrossRef]
- Clendenen, L.H. Intra- and Inter-Operator Reliability of Cephalometric Landmark Identification: A Comparison Amongst CBCT Ray-Sum, Maximum Intensity Projection Techniques, and Conventional Digital Cephalometric Radiographs; University of Missouri—Kansas City: Kansas City, MO, USA, 2010. [Google Scholar]
- Lee, J.G.; Jun, S.; Cho, Y.W.; Lee, H.; Kim, G.B.; Seo, J.B. Kim, N. Deep Learning in Medical Imaging: General Overview. Korean J. Radiol. 2017, 18, 570–584. [Google Scholar] [CrossRef]
- Dorgham, O.M.; Laycock, S.D.; Fisher, M.H. GPU accelerated generation of digitally reconstructed radiographs for 2-D/3-D image registration. IEEE Trans. Biomed. Eng. 2012, 59, 594–603. [Google Scholar] [CrossRef]
- Ruellas, A.C.; Tonello, C.; Gomes, L.R.; Yatabe, M.S.; Macron, L.; Lopinto, J.; Goncalves, J.R.; Garib Carreira, D.G.; Alonso, N.; Souki, B.Q.; et al. Common 3-dimensional coordinate system for assessment of directional changes. Am. J. Orthod. Dentofacial. Orthop. 2016, 149, 645–656. [Google Scholar] [CrossRef]
- Jung, P.G.; Lee, G.C.; Moon, C.H. Comparison of cone-beam computed tomography cephalometric measurements using a midsagittal projection and conventional two-dimensional cephalometric measurements. Korean J. Orthod. 2015, 45, 282–288. [Google Scholar] [CrossRef]
- Lee, E.H.; Yu, H.S.; Lee, K.J.; Han, S.S.; Jung, H.D. Comparison of three midsagittal planes for three-dimensional cone beam computed tomography head reorientation. Korean J. Orthod. 2020, 50, 3–12. [Google Scholar] [CrossRef]
- Cattaneo, P.M.; Melsen, B. The use of cone-beam computed tomography in an orthodontic department in between research and daily clinic. World J. Orthod. 2008, 9, 269–282. [Google Scholar] [PubMed]
- Levy-Mandel, A.D.; Venetsamopolus, A.N.; Tsosos, J.K. Knowledge based landmarking of cephalograms. Comput. Biomed. Res. 1986, 19, 282–309. [Google Scholar] [CrossRef]
- Ibragimov, B.; Likar, B.; Pernuš, F.; Tomaž, V. Automatic Cephalometric X-Ray Landmark Detection by Applying Game Theory and Random Forests. Proc. ISBI Int. Symp. Biomedical. Imaging 2014. [Google Scholar]
- Leonardi, R.; Giordano, D.; Maiorana, F.; Spampinato, C. Automatic Cephalometric Analysis: A Systematic Review. Angle Orthod. 2008, 78, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Lindner, C.; Wang, C.W.; Huang, C.T.; Li, C.H.; Chang, S.W.; Cootes, T.F. Fully Automatic System for Accurate Localisation and Analysis of Cephalometric Landmarks in Lateral Cephalograms. Sci. Rep. 2016, 6, 33581. [Google Scholar] [CrossRef]
- Wang, S.; Li, H.; Li, J.; Zhang, Y.; Zou, B. Automatic Analysis of Lateral Cephalograms Based on Multiresolution Decision Tree Regression Voting. J. Healthc. Eng. 2018, 2018, 1797502. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural. Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69S, S36–S40. [Google Scholar] [CrossRef]
- Zhang, Y.; An, M. Deep Learning- and Transfer Learning-Based Super Resolution Reconstruction from Single Medical Image. J. Healthc. Eng. 2017, 2017, 5859727. [Google Scholar] [CrossRef]
- Arık, S.Ö.; Ibragimov, B.; Xing, L. Fully automated quantitative cephalometry using convolutional neural networks. J. Med Imaging 2017, 4, 014501. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, S.; Sotsuka, Y.; Kawai, K.; Ishise, H.; Kakibuchi, M. Personal Computer-Based Cephalometric Landmark Detection With Deep Learning, Using Cephalograms on the Internet. J. Craniofac. Surg. 2019, 30, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Hwang, H.W.; Moon, J.H.; Yu, Y.S.; Kim, H.S.; Her, S.B.; Srinivasan, G.; Aljanabi, M.N.A.; Donatelli, R.E.; Lee, S.J. Automated identification of cephalometric landmarks: Part 1—Comparisons between the latest deep-learning methods YOLOV3 and SSD. Angle Orthod. 2019, 89, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Liu, Y.; Oh, S.H.; Ahn, H.W.; Kim, S.H.; Nelson, G. Evaluation of multi-stage convolutional neural network based fully automatic landmark identification system using CBCT synthesized posteroanterior cephalometric images. Korean J. Orthod. 2020; in press. [Google Scholar]
- Lee, Y.J.; Park, J.H.; Chang, N.Y.; Lee, M.Y.; Kim, B.C.; Seo, H.Y.; Mangal, U.; Chae, J.M. Assessment of bone density changes following two-jaw surgery using multidetector computed tomography: A pilot study. Korean J. Orthod. 2020, 50, 157–169. [Google Scholar] [CrossRef]
- Cho, J.H.; Lee, K.W.; Shin, E. How Much Data Is Needed to Train A Medical Image Deep Learning System to Achieve Necessary High Accuracy. arXiv 2015, 1511, 06348. [Google Scholar]
- Steyerberg, E.W.; Eijkemans, M.J.C.; Harrell, F.E.; Habbema, J.; Dik, F. Prognostic Modeling with Logistic Regression Analysis: In Search of a Sensible Strategy in Small Data Sets. Med Decis. Mak. 2001, 21, 45–56. [Google Scholar] [CrossRef]
- Kim, S.H.; Ahn, H.W.; Chung, K.R.; Gerald, N. The biocreative strategy. Part 2: The Tweemac analysis. J. Clin. Orthod. 2018, 52, 359–371. [Google Scholar]
- Hwang, H.W.; Park, J.H.; Moon, J.H.; Yu, Y.S.; Kim, H.S.; Her, S.B.; Srinivasa, G.; Aljanabi, M.N.A.; Donatelli, R.E.; Lee, S.J. Automated Identification of Cephalometric Landmarks: Part 2-Might It Be Better Than human? Angle Orthod. 2019, 90, 69–76. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J.M. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging. 2016, 35, 1299–1312. [Google Scholar] [CrossRef]
- Anwar, S.H.; Majid, M.; Qayyum, A.; Awais, M.; Alnowam, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef]
- Schwendicke, F.; Golla, T.; Dreher, M.; Krois, J. Convolutional neural networks for dental image diagnostics: A scoping review. J. Dent. 2019, 91, 103226. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big. Data 2019, 6, 113. [Google Scholar] [CrossRef]
- Takahashiy, R.; Takashi, M.; Kuniaki. Multi-Stage Convolutional Neural Networks for Robustness to Scale Transformation. Int. Symp. Nonlinear Theory Appl. 2017. [Google Scholar] [CrossRef]
- Takahashi, R.; Matsubara, T.; Uehara, K. A Novel Weight-Shared Multi-Stage CNN for Scale Robustness. IEEE Trans. Circuits Syst. Video Technol. 2019, 29, 1090–1101. [Google Scholar] [CrossRef]
- Sun, M.L.; Song, Z.J.; Jiang, X.H.; Pan, J.; Pang, Y.W. Learning Pooling for Convolutional Neural Network. Neurocomputing 2017, 224, 96–104. [Google Scholar] [CrossRef]
- Wallis, J.W.; Miller, T.R.; Lerner, C.A.; Kleerup, E.C. Three-dimensional display in nuclear medicine. IEEE Trans. Med. Imaging 1989, 8, 297–300. [Google Scholar] [CrossRef]
- Wallis, J.W.; Miller, T.R. Three-dimensional display in nuclear medicine and radiology. J. Nucl. Med. 1991, 32, 534–546. [Google Scholar]
- Rossini, G.; Cavallini, C.; Cassetta, M. 3D cephalometric analysis obtained from computed tomography. Review of the literature. Ann. Stomatol. 2011, 2, 31–39. [Google Scholar]
- Navarro, R.L.; Oltramari-Navarro, P.V.P.; Fernandes, T.M.F.; Oliveira, G.F.; Conti, A.C.C.; Almeida, M.R.; Almeida, R.R. Comparison of manual, digital and lateral CBCT cephalometric analyses. J. Appl. Oral Sci. 2013, 21, 167–176. [Google Scholar] [CrossRef]
- Gribel, B.F.; Gribel, M.N.; Manzi, F.R.; Brooks, S.L.; McNamara, J.A., Jr. From 2D to 3D: An algorithm to derive normal values for 3-dimensional computerized assessment. Angle Orthod. 2011, 81, 3–10. [Google Scholar] [CrossRef]
- Park, C.S.; Park, J.K.; Kim, H.; Han, S.S.; Jeong, H.G.; Park, H. Comparison of conventional lateral cephalograms with corresponding CBCT radiographs. Imaging Sci. Dent. 2012, 42, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Kunz, F.; Stellzig-Eisenhauer, A.; Zeman, F.; Boldt, J. Artificial intelligence in orthodontics. J. Orofac. Orthop. 2020, 81, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.N.; Shim, E.J.; Park, J.E.; Kim, Y.J.; Lee, U.Y.; Kim, Y.J. Web-based fully automated cephalometric analysis by deep learning. Comput. Methods Programs Biomed. 2020, 194, 105513. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yu, H.J.; Kim, M.J.; Kim, J.W.; Choi, J.E. Automated cephalometric landmark detection with confidence regions using Bayesian convolutional neural networks. BMC Oral Health 2020, 20, 270. [Google Scholar] [CrossRef]
- Moon, J.H.; Hwang, H.W.; Yu, Y.S.; Kim, M.G.; Donatelli, R.E.; Lee, S.J. How much deep learning is enough for automatic identification to be reliable?: A cephalometric example. Angle Orthod. 2020, 90, 823–830. [Google Scholar] [CrossRef]






| Landmarks | Definition |
|---|---|
| Nasion (N) | The most anterior point of the frontonasal suture |
| Orbitale (Or) | Inferior margin of the orbit |
| Porion (Po) | Superior margin of the external auditory meatus |
| Anterior nasal spine (ANS) | Tip of anterior nasal spine |
| A point | Point at the deepest concavity on the maxilla between the anterior nasal spine and prosthion |
| Maxillary central incisor root (Mx1r) | Maxillary central incisor root |
| Maxillary incisal edge (Is) | Maxillary incisal edge |
| Mandibular incisal edge (Ii) | Mandibular incisal edge |
| Infradentale (Id) | The highest and most anterior point on the alveolar process in the median plane between the mandibular central incisors |
| Mandibular central incisor root (Md1r) | Mandibular central incisor root |
| B point | Point at the deepest concavity on the mandibular symphysis between infradentale and pogonion |
| Pogonion (Pog) | The most anterior midpoint of the chin of the mandibular symphysis |
| Menton (Me) | The most inferior point of the mandibular symphysis |
| Gonion (Go) | Most posterior inferior point on angle of mandible |
| Posterior occlusal plane point (Pop) | Posterior occlusal plane point: mesio-buccal cusp of 1st molar |
| Landmark. | MRE (mm) | SD (mm) | SDR (%) | |||
|---|---|---|---|---|---|---|
| 2.0 mm | 2.5 mm | 3.0 mm | 4.0 mm | |||
| N | 0.56 | 1.497 | 95.97 | 96.55 | 97.12 | 98.27 |
| Or | 0.69 | 1.397 | 97.12 | 98.27 | 98.27 | 98.85 |
| Po | 1.43 | 1.689 | 78.73 | 84.48 | 89.65 | 94.82 |
| ANS | 0.76 | 0.726 | 93.67 | 97.12 | 98.27 | 99.42 |
| A | 1.22 | 2.697 | 89.08 | 91.95 | 96.55 | 98.27 |
| Mx1r | 1.59 | 1.308 | 69.54 | 78.73 | 86.2 | 95.4 |
| Is | 0.61 | 1.081 | 93.1 | 94.25 | 94.82 | 98.27 |
| Id | 0.79 | 1.080 | 90.8 | 94.25 | 95.4 | 96.55 |
| Ii | 0.65 | 0.927 | 93.67 | 94.25 | 95.97 | 97.7 |
| Md1r | 1.39 | 1.266 | 81.03 | 88.5 | 89.65 | 94.25 |
| B | 1.09 | 0.891 | 83.33 | 92.52 | 95.97 | 98.85 |
| Pog | 0.58 | 0.580 | 95.97 | 97.7 | 98.85 | 100 |
| Me | 0.59 | 0.547 | 97.12 | 98.27 | 98.85 | 100 |
| Go | 2.04 | 1.727 | 62.64 | 72.98 | 77.58 | 85.63 |
| Pop | 1.31 | 1.897 | 85.05 | 87.93 | 89.65 | 92.52 |
| Average | 1.03 | 1.288 | 87.13 | 91.19 | 93.52 | 96.59 |
| Truth Ground | AI Prediction CBCT-LC | p-Value * | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Landmark | Mean | SD | Min | Max | Mean | SD | Min | Max | |
| N_x | 1642.71 | 58.46 | 1479.00 | 1866.00 | 1641.33 | 55.81 | 1487.66 | 1849.16 | 0.868 |
| N_y | 564.09 | 124.36 | 285.00 | 865.00 | 551.58 | 122.73 | 259.93 | 828.51 | 0.488 |
| Or_x | 1561.92 | 55.80 | 1363.00 | 1755.00 | 1558.83 | 54.22 | 1378.06 | 1732.80 | 0.703 |
| Or_y | 819.48 | 105.41 | 628.00 | 1079.00 | 805.06 | 105.67 | 604.01 | 1047.61 | 0.350 |
| Po_x | 851.02 | 81.24 | 526.00 | 991.00 | 850.27 | 78.83 | 530.84 | 989.41 | 0.950 |
| Po_y | 827.38 | 107.98 | 633.00 | 1085.00 | 819.01 | 107.73 | 612.38 | 1088.11 | 0.593 |
| ANS_x | 1665.24 | 55.63 | 1505.00 | 1875.00 | 1663.51 | 52.62 | 1497.87 | 1838.49 | 0.829 |
| ANS_y | 1052.13 | 86.66 | 883.00 | 1280.00 | 1042.43 | 90.04 | 834.11 | 1275.94 | 0.453 |
| A_x | 1642.36 | 54.26 | 1496.00 | 1851.00 | 1640.73 | 52.12 | 1497.96 | 1831.31 | 0.833 |
| A_y | 1105.75 | 84.83 | 946.00 | 1327.00 | 1095.83 | 88.24 | 914.74 | 1325.59 | 0.423 |
| Mx1r_x | 1618.64 | 53.64 | 1468.00 | 1813.00 | 1612.80 | 50.99 | 1479.12 | 1779.62 | 0.457 |
| Mx1r_y | 1134.80 | 79.14 | 969.00 | 1349.00 | 1128.02 | 79.84 | 951.91 | 1323.55 | 0.545 |
| Is_x | 1697.26 | 68.22 | 1534.00 | 1926.00 | 1697.33 | 65.10 | 1543.72 | 1899.46 | 0.994 |
| Is_y | 1314.39 | 75.45 | 1150.00 | 1499.00 | 1306.30 | 78.15 | 1123.21 | 1506.45 | 0.455 |
| Id_x | 1655.10 | 79.85 | 1475.00 | 1873.00 | 1650.89 | 75.27 | 1482.44 | 1848.20 | 0.716 |
| Id_y | 1392.60 | 64.48 | 1262.00 | 1546.00 | 1384.93 | 70.47 | 1224.56 | 1557.32 | 0.401 |
| Ii_x | 1677.85 | 75.42 | 1515.00 | 1902.00 | 1677.02 | 70.94 | 1514.51 | 1875.32 | 0.939 |
| Ii_y | 1305.05 | 69.12 | 1164.00 | 1475.00 | 1294.47 | 72.27 | 1129.55 | 1466.30 | 0.282 |
| Md1r_x | 1596.45 | 82.52 | 1414.00 | 1788.00 | 1590.17 | 78.16 | 1415.73 | 1778.64 | 0.605 |
| Md1r_y | 1453.87 | 58.29 | 1336.00 | 1575.00 | 1445.50 | 64.19 | 1283.63 | 1621.69 | 0.326 |
| B_x | 1607.09 | 86.88 | 1424.00 | 1813.00 | 1602.59 | 82.05 | 1424.10 | 1785.40 | 0.723 |
| B_y | 1506.37 | 64.14 | 1337.00 | 1628.00 | 1497.51 | 71.16 | 1294.64 | 1650.67 | 0.358 |
| Pog_x | 1613.85 | 93.19 | 1419.00 | 1837.00 | 1610.07 | 88.90 | 1419.37 | 1809.18 | 0.785 |
| Pog_y | 1601.34 | 58.20 | 1471.00 | 1704.00 | 1600.11 | 58.44 | 1442.15 | 1712.63 | 0.880 |
| Me_x | 1555.66 | 96.74 | 1352.00 | 1783.00 | 1553.77 | 91.11 | 1355.66 | 1748.78 | 0.894 |
| Me_y | 1663.93 | 55.23 | 1526.00 | 1770.00 | 1658.06 | 55.99 | 1510.33 | 1773.85 | 0.445 |
| Go_x | 992.75 | 87.06 | 725.00 | 1184.00 | 988.62 | 79.60 | 719.04 | 1152.65 | 0.748 |
| Go_y | 1373.97 | 65.90 | 1202.00 | 1537.00 | 1355.27 | 64.39 | 1213.18 | 1528.06 | 0.053 |
| Pop_x | 1434.15 | 58.52 | 1264.00 | 1616.00 | 1425.49 | 56.55 | 1261.20 | 1588.02 | 0.314 |
| Pop_y | 1263.29 | 65.40 | 1130.00 | 1426.00 | 1253.58 | 68.57 | 1104.16 | 1427.76 | 0.292 |
| Truth Ground | AI prediction MIP-LC | p-Value * | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Landmark | Mean | SD | Min | Max | Mean | SD | Min | Max | |
| N_x | 1642.71 | 58.46 | 1479.00 | 1866.00 | 1646.17 | 59.46 | 1479.27 | 1862.75 | 0.754 |
| N_y | 564.09 | 124.36 | 285.00 | 865.00 | 562.05 | 118.36 | 288.38 | 830.26 | 0.793 |
| Or_x | 1561.92 | 55.80 | 1363.00 | 1755.00 | 1565.48 | 57.18 | 1369.46 | 1756.59 | 0.649 |
| Or_y | 819.48 | 105.41 | 628.00 | 1079.00 | 819.07 | 100.80 | 630.06 | 1048.80 | 0.959 |
| Po_x | 851.02 | 81.24 | 526.00 | 991.00 | 850.48 | 81.05 | 517.92 | 986.90 | 0.920 |
| Po_y | 827.38 | 107.98 | 633.00 | 1085.00 | 822.26 | 101.70 | 635.18 | 1053.43 | 0.156 |
| ANS_x | 1665.24 | 55.63 | 1505.00 | 1875.00 | 1670.20 | 56.72 | 1505.48 | 1875.80 | 0.406 |
| ANS_y | 1052.13 | 86.66 | 883.00 | 1280.00 | 1052.22 | 86.83 | 875.05 | 1278.58 | 0.752 |
| A_x | 1642.36 | 54.26 | 1496.00 | 1851.00 | 1645.53 | 55.47 | 1496.60 | 1849.64 | 0.778 |
| A_y | 1105.75 | 84.83 | 946.00 | 1327.00 | 1107.52 | 83.06 | 950.80 | 1328.57 | 0.752 |
| Mx1r_x | 1618.64 | 53.64 | 1468.00 | 1813.00 | 1621.62 | 54.20 | 1479.40 | 1825.12 | 0.771 |
| Mx1r_y | 1134.80 | 79.14 | 969.00 | 1349.00 | 1138.54 | 77.21 | 980.84 | 1351.76 | 0.333 |
| Is_x | 1697.26 | 68.22 | 1534.00 | 1926.00 | 1701.67 | 68.60 | 1542.86 | 1927.34 | 0.601 |
| Is_y | 1314.39 | 75.45 | 1150.00 | 1499.00 | 1314.79 | 74.57 | 1149.57 | 1501.82 | 0.599 |
| Id_x | 1655.10 | 79.85 | 1475.00 | 1873.00 | 1655.84 | 78.72 | 1482.70 | 1873.20 | 0.799 |
| Id_y | 1392.60 | 64.48 | 1262.00 | 1546.00 | 1397.29 | 65.69 | 1262.71 | 1561.40 | 0.381 |
| Ii_x | 1677.85 | 75.42 | 1515.00 | 1902.00 | 1679.90 | 74.65 | 1512.09 | 1899.68 | 0.986 |
| Ii_y | 1305.05 | 69.12 | 1164.00 | 1475.00 | 1307.15 | 69.11 | 1164.50 | 1474.31 | 0.770 |
| Md1r_x | 1596.45 | 82.52 | 1414.00 | 1788.00 | 1596.19 | 82.44 | 1408.86 | 1815.76 | 0.684 |
| Md1r_y | 1453.87 | 58.29 | 1336.00 | 1575.00 | 1456.71 | 62.78 | 1310.59 | 1610.44 | 0.997 |
| B_x | 1607.09 | 86.88 | 1424.00 | 1813.00 | 1608.83 | 85.34 | 1422.78 | 1812.24 | 0.979 |
| B_y | 1506.37 | 64.14 | 1337.00 | 1628.00 | 1507.28 | 67.61 | 1326.61 | 1642.09 | 0.435 |
| Pog_x | 1613.85 | 93.19 | 1419.00 | 1837.00 | 1615.02 | 91.87 | 1415.98 | 1837.82 | 0.915 |
| Pog_y | 1601.34 | 58.20 | 1471.00 | 1704.00 | 1607.11 | 57.09 | 1467.66 | 1703.10 | 0.294 |
| Me_x | 1555.66 | 96.74 | 1352.00 | 1783.00 | 1556.17 | 94.12 | 1353.87 | 1767.15 | 0.862 |
| Me_y | 1663.93 | 55.23 | 1526.00 | 1770.00 | 1666.42 | 55.77 | 1522.65 | 1768.89 | 0.657 |
| Go_x | 992.75 | 87.06 | 725.00 | 1184.00 | 992.71 | 81.71 | 719.39 | 1169.11 | 0.941 |
| Go_y | 1373.97 | 65.90 | 1202.00 | 1537.00 | 1374.04 | 63.35 | 1186.13 | 1531.91 | 0.493 |
| Pop_x | 1434.15 | 58.52 | 1264.00 | 1616.00 | 1429.53 | 60.57 | 1253.62 | 1616.57 | 0.064 |
| Pop_y | 1263.29 | 65.40 | 1130.00 | 1426.00 | 1261.56 | 65.56 | 1131.64 | 1427.02 | 0.182 |
| Method. | Landmark | Total Data | SDR (%) | |||
|---|---|---|---|---|---|---|
| 2.0 mm | 2.5 mm | 3.0 mm | 4.0 mm | |||
| SDD22 | 19 | 400 | 75.6 | 81.3 | 84.7 | 88.1 |
| YOLOv324 | 80 | 1311 | 80.4 | 87.4 | 92.0 | 96.2 |
| Web-based45 | 23 | 2075 | 84.5 | 90.1 | 93.2 | 96.8 |
| Proposed | 15 | 860 | 87.1 | 91.2 | 93.5 | 96.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.-J.; Liu, Y.; Oh, S.H.; Ahn, H.-W.; Kim, S.-H.; Nelson, G. Automatic Cephalometric Landmark Identification System Based on the Multi-Stage Convolutional Neural Networks with CBCT Combination Images. Sensors 2021, 21, 505. https://doi.org/10.3390/s21020505
Kim M-J, Liu Y, Oh SH, Ahn H-W, Kim S-H, Nelson G. Automatic Cephalometric Landmark Identification System Based on the Multi-Stage Convolutional Neural Networks with CBCT Combination Images. Sensors. 2021; 21(2):505. https://doi.org/10.3390/s21020505
Chicago/Turabian StyleKim, Min-Jung, Yi Liu, Song Hee Oh, Hyo-Won Ahn, Seong-Hun Kim, and Gerald Nelson. 2021. "Automatic Cephalometric Landmark Identification System Based on the Multi-Stage Convolutional Neural Networks with CBCT Combination Images" Sensors 21, no. 2: 505. https://doi.org/10.3390/s21020505
APA StyleKim, M.-J., Liu, Y., Oh, S. H., Ahn, H.-W., Kim, S.-H., & Nelson, G. (2021). Automatic Cephalometric Landmark Identification System Based on the Multi-Stage Convolutional Neural Networks with CBCT Combination Images. Sensors, 21(2), 505. https://doi.org/10.3390/s21020505






