Video-Rate Quantitative Phase Imaging Using a Digital Holographic Microscope and a Generative Adversarial Network
Abstract
:1. Introduction
2. Materials and Methods
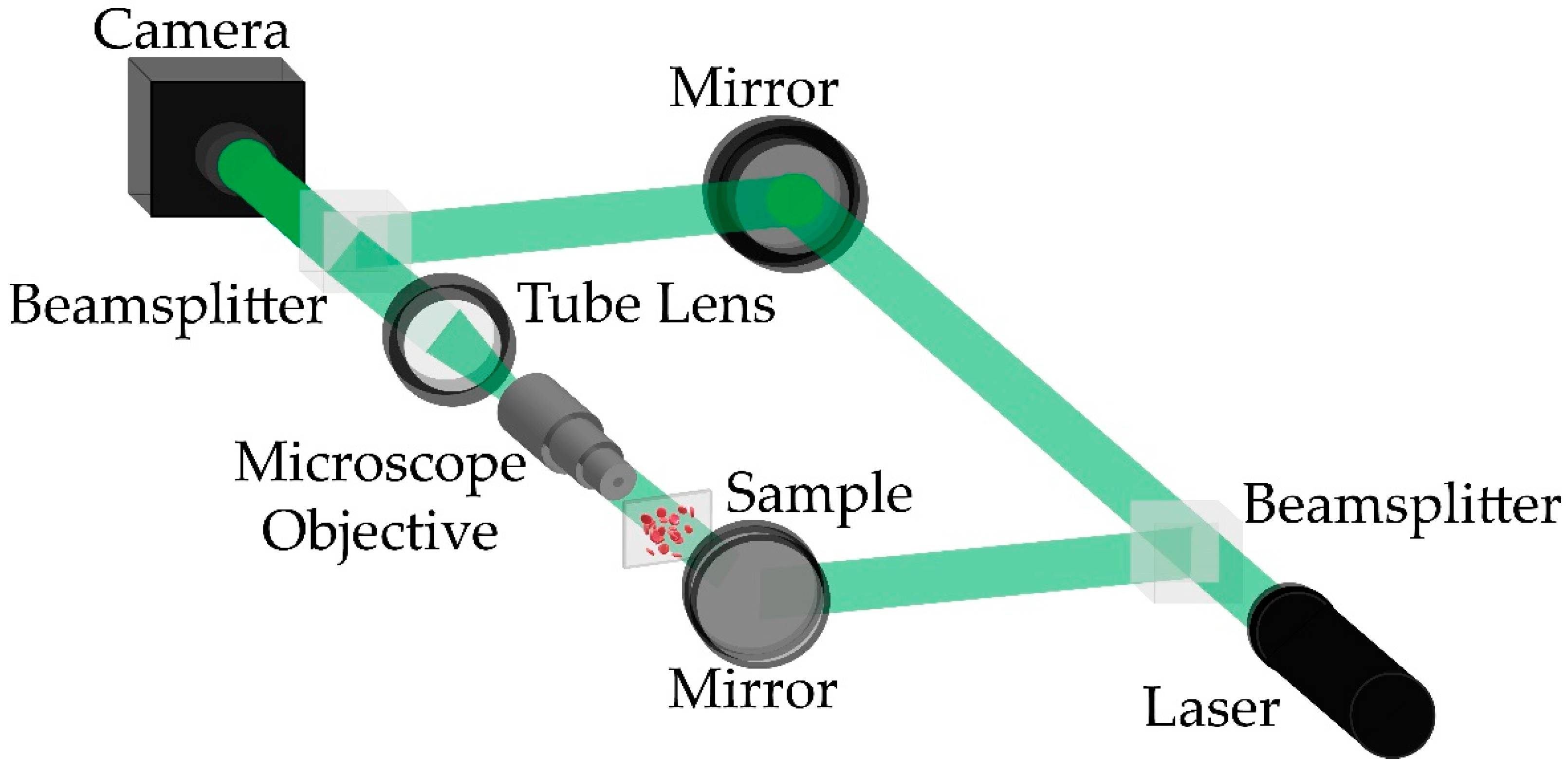
2.1. Experimental System
2.2. The Dataset
2.3. The Proposed Learning-Based Method
3. Experimental Results
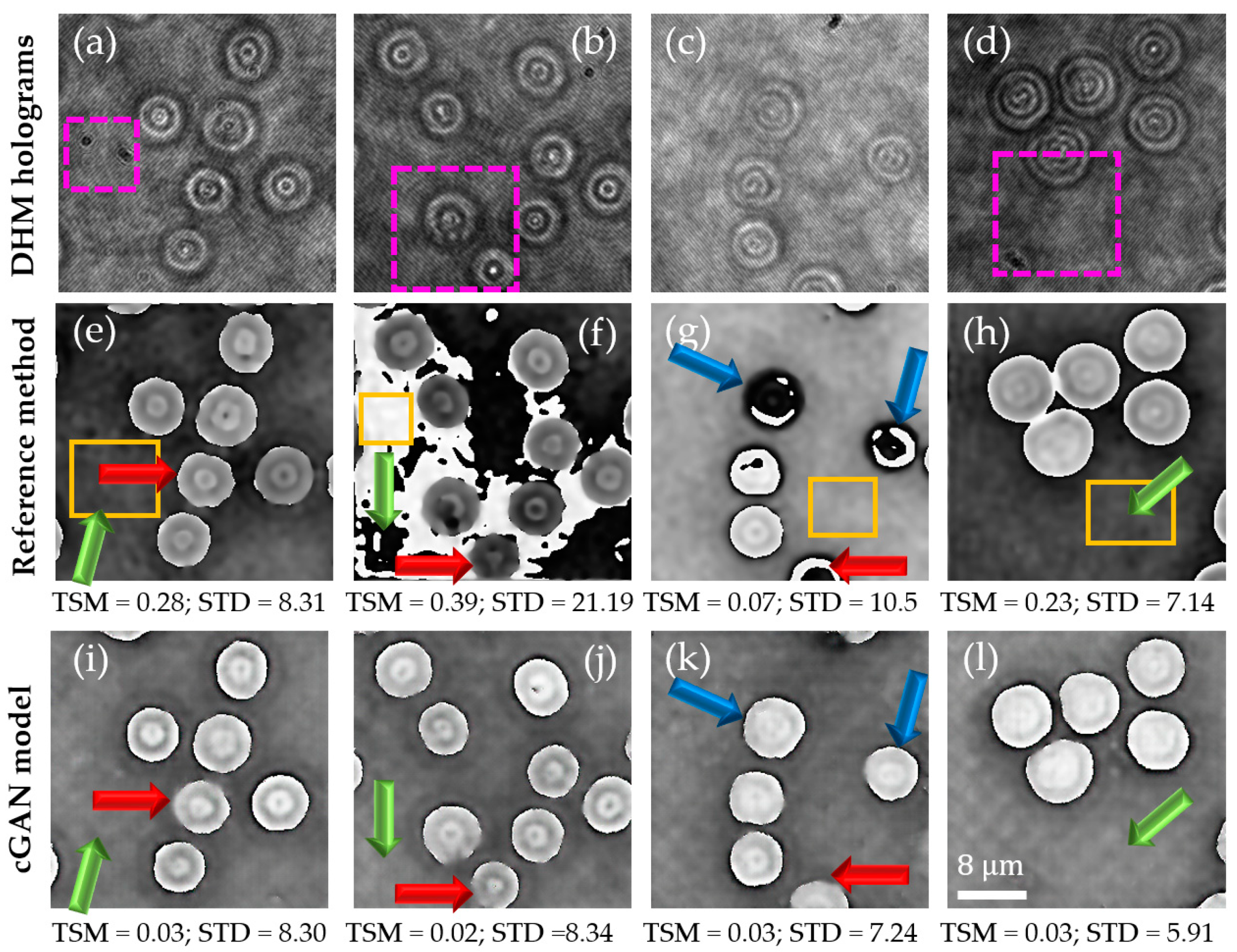
3.1. Experimental Quantitative Phase Images Obtained by the Proposed Learning-Based Model Using Static DHM Holograms
3.2. Comparison of the Proposed Cgan Model against the U-Net Model and Validation of the Proposal’s Generalization Ability to System’s Diversity
3.3. Validation of the Proposed Learning-Based Model Using a Sequence of Dynamic DHM Holograms for Video-Rate Quantitative Phase Images
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cacace, T.; Bianco, V.; Ferraro, P. Quantitative phase imaging trends in biomedical applications. Opt. Lasers Eng. 2020, 135, 106188. [Google Scholar] [CrossRef]
- Popescu, G. Quantitative Phase Imaging of Cells and Tissues; McGraw-Hill: New York, NY, USA, 2012; Volume 17, ISBN 978-0-07-166342-7. [Google Scholar]
- Park, Y.K.; Depeursinge, C.; Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photonics 2018, 12, 578–589. [Google Scholar] [CrossRef]
- Park, Y.; Popescu, G.; Ferraro, P.; Kemper, B. Editorial: Quantitative Phase Imaging and Its Applications to Biophysics, Biology, and Medicine. Front. Phys. 2020, 7, 2019–2020. [Google Scholar] [CrossRef] [Green Version]
- Gureyev, T.E.; Nugent, K.A. Rapid quantitative phase imaging using the transport of intensity equation. Opt. Commun. 1997, 133, 339–346. [Google Scholar] [CrossRef]
- Tian, X.; Yu, W.; Meng, X.; Sun, A.; Xue, L.; Liu, C.; Wang, S. Real-time quantitative phase imaging based on transport of intensity equation with dual simultaneously recorded field of view. Opt. Lett. 2016, 41, 1427. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.; Bhaduri, B.; Wang, R.; Zhu, R.; Popescu, G. Quantitative Phase Imaging; Elsevier Inc.: Amsterdam, The Netherlands, 2012; Volume 57, ISBN 9780444594228. [Google Scholar]
- Trusiak, M.; Mico, V.; Garcia, J.; Patorski, K. Quantitative phase imaging by single-shot Hilbert–Huang phase microscopy. Opt. Lett. 2016, 41, 4344. [Google Scholar] [CrossRef] [PubMed]
- Trusiak, M.; Cywinska, M.; Mico, V.; Picazo-Bueno, J.A.; Zuo, C.; Zdankowski, P.; Patorski, K. Variational Hilbert Quantitative Phase Imaging. Sci. Rep. 2020, 10, 13955. [Google Scholar] [CrossRef]
- Trimby, P.W.; Prior, D.J. Microstructural imaging techniques: A comparison between light and scanning electron microscopy. Tectonophysics 1999, 303, 71–81. [Google Scholar] [CrossRef]
- Stagaman, G.; Forsyth, J.M. Bright-field microscopy of semitransparent objects. J. Opt. Soc. Am. A 1988, 5, 648. [Google Scholar] [CrossRef]
- Guo, C.-S.; Wang, B.-Y.; Sha, B.; Lu, Y.-J.; Xu, M.-Y. Phase derivative method for reconstruction of slightly off-axis digital holograms. Opt. Express 2014, 22, 30553. [Google Scholar] [CrossRef]
- Javidi, B.; Carnicer, A.; Anand, A.; Barbastathis, G.; Chen, W.; Ferraro, P.; Goodman, J.W.; Horisaki, R.; Khare, K.; Kujawinska, M.; et al. Roadmap on digital holography. Opt. Express 2021, 29, 35078–35118. [Google Scholar] [CrossRef]
- Kim, M.K. Principles and techniques of digital holographic microscopy. SPIE Rev. 2010, 1, 18005. [Google Scholar] [CrossRef] [Green Version]
- Cheong, F.C.; Krishnatreya, B.J.; Grier, D.G. Strategies for three-dimensional particle tracking with holographic video microscopy. Opt. Express 2010, 18, 13563–13573. [Google Scholar] [CrossRef]
- Yu, X.; Hong, J.; Liu, C.; Kim, M.K. Review of digital holographic microscopy for three-dimensional profiling and tracking. Opt. Eng. 2014, 53, 112306. [Google Scholar] [CrossRef] [Green Version]
- Dubois, F.; Yourassowsky, C.; Monnom, O.; Legros, J.-C.; Debeir, O.; Van Ham, P.; Kiss, R.; Decaestecker, C. Digital holographic microscopy for the three-dimensional dynamic analysis of in vitro cancer cell migration. J. Biomed. Opt. 2006, 11, 054032. [Google Scholar] [CrossRef]
- O’Connor, T.; Anand, A.; Andemariam, B.; Javidi, B. Overview of cell motility-based sickle cell disease diagnostic system in shearing digital holographic microscopy. J. Phys. Photonics 2020, 2, 031002. [Google Scholar] [CrossRef] [Green Version]
- Hellesvik, M.; Øye, H.; Aksnes, H. Exploiting the potential of commercial digital holographic microscopy by combining it with 3D matrix cell culture assays. Sci. Rep. 2020, 10, 14680. [Google Scholar] [CrossRef]
- Kemper, B.; Carl, D.; Schnekenburger, J.; Bredebusch, I.; Schäfer, M.; Domschke, W.; von Bally, G. Investigation of living pancreas tumor cells by digital holographic microscopy. J. Biomed. Opt. 2006, 11, 034005. [Google Scholar] [CrossRef] [PubMed]
- Montfort, F.; Emery, Y.; Marquet, F.; Cuche, E.; Aspert, N.; Solanas, E.; Mehdaoui, A.; Ionescu, A.; Depeursinge, C. Process engineering and failure analysis of MEMS and MOEMS by digital holography microscopy (DHM). Proc. SPIE 2007, 6463, 64630G. [Google Scholar]
- Kim, M.K. Applications of digital holography in biomedical microscopy. J. Opt. Soc. Korea 2010, 14, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Castañeda, R.; Garcia-Sucerquia, J. Single-shot 3D topography of reflective samples with digital holographic microscopy. Appl. Opt. 2018, 57, A12. [Google Scholar] [CrossRef]
- Osten, W. Digital Holography and Its Application in MEMS/MOEMS Inspection, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2019; ISBN 9780429186738. [Google Scholar]
- Doblas, A.; Sánchez-Ortiga, E.; Martínez-Corral, M.; Saavedra, G.; Garcia-Sucerquia, J. Accurate single-shot quantitative phase imaging of biological specimens with telecentric digital holographic microscopy. J. Biomed. Opt. 2014, 19, 46022. [Google Scholar] [CrossRef] [Green Version]
- Trujillo, C.; Castañeda, R.; Piedrahita-Quintero, P.; Garcia-Sucerquia, J. Automatic full compensation of quantitative phase imaging in off-axis digital holographic microscopy. Appl. Opt. 2016, 55, 10299–10306. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Nguyen, C.V.; Pratap, M.; Zheng, Y.; Wang, Y.; Nisbet, D.R.; Williams, R.J.; Rug, M.; Maier, A.G.; Lee, W.M. Automated Fourier space region-recognition filtering for off-axis digital holographic microscopy. Biomed. Opt. Express 2016, 7, 3111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.; Bui, V.; Lam, V.; Raub, C.B.; Chang, L.-C.; Nehmetallah, G. Automatic phase aberration compensation for digital holographic microscopy based on deep learning background detection. Opt. Express 2017, 25, 15043–15057. [Google Scholar] [CrossRef] [PubMed]
- Rivenson, Y.; Zhang, Y.; Günaydın, H.; Teng, D.; Ozcan, A. Phase recovery and holographic image reconstruction using deep learning in neural networks. Light Sci. Appl. 2018, 7, 17141. [Google Scholar] [CrossRef]
- Anand, A.; Chhaniwal, V.K.; Javidi, B. Real-time digital holographic microscopy for phase contrast 3D imaging of dynamic phenomena. IEEE/OSA J. Disp. Technol. 2010, 6, 500–505. [Google Scholar] [CrossRef]
- Pitkäaho, T.; Manninen, A.; Naughton, T.J. Focus prediction in digital holographic microscopy using deep convolutional neural networks. Appl. Opt. 2019, 58, A202–A208. [Google Scholar]
- Pitkäaho, T.; Manninen, A.; Naughton, T.J. Deep convolutional neural networks and digital holographic microscopy for in-focus depth estimation of microscopic objects. In Proceedings of the Irish Machine Vision and Image Processing Conference Proceedings, Maynooth, Ireland, 30 August–1 September 2017; pp. 52–59. [Google Scholar]
- Liu, T.; Wei, Z.; Rivenson, Y.; de Haan, K.; Zhang, Y.; Wu, Y.; Ozcan, A. Color Holographic Microscopy Using a Deep Neural Network. In Proceedings of the Conference on Lasers and Electro-Optics, Optical Society of America, Washington, DC, USA, 10–15 May 2020; p. 1.AM1I.1. [Google Scholar]
- Yin, D.; Gu, Z.; Zhang, Y.; Gu, F.; Nie, S.; Ma, J.; Yuan, C. Digital holographic reconstruction based on deep learning framework with unpaired data. IEEE Photonics J. 2020, 12, 3900312. [Google Scholar] [CrossRef]
- Rivenson, Y.; Wu, Y.; Ozcan, A. Deep learning in holography and coherent imaging. Light Sci. Appl. 2019, 8, 85. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Dou, J.; Kemao, Q.; Di, J.; Zhao, J. Y-Net: A one-to-two deep learning framework for digital holographic reconstruction. Opt. Lett. 2019, 44, 4765–4768. [Google Scholar] [CrossRef] [PubMed]
- Vijayanagaram, R. Application of Deep Learning Techniques to Digital Holographic Microscopy for Numerical Reconstruction. In Proceedings of theAll-Russian Conference “Spatial Data Processing for Monitoring of Natural and Anthropogenic Processes” (SDM-2019), Berdsk, Russia, 26–30 August 2019; CEUR-WS: Aachen, Germany, 2020; Volume 2535, pp. 1–12. [Google Scholar]
- Di, J.; Wu, J.; Wang, K.; Tang, J.; Li, Y.; Zhao, J. Quantitative Phase Imaging Using Deep Learning-Based Holographic Microscope. Front. Phys. 2021, 9, 1–7. [Google Scholar] [CrossRef]
- Moon, I.; Jaferzadeh, K.; Kim, Y.; Javidi, B. Noise-free quantitative phase imaging in Gabor holography with conditional generative adversarial network. Opt. Express 2020, 28, 26284–26301. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Liu, Q.; Yu, Y.; Luo, Y.; Wang, S. Quantitative phase imaging in digital holographic microscopy based on image inpainting using a two-stage generative adversarial network. Opt. Express 2021, 29, 24928–24946. [Google Scholar] [CrossRef] [PubMed]
- Cuche, E.; Marquet, P.; Depeursinge, C. Simultaneous amplitude-contrast and quantitative phase-contrast microscopy by numerical reconstruction of Fresnel off-axis holograms. Appl. Opt. 1999, 38, 6994–7001. [Google Scholar] [CrossRef] [PubMed]
- Colomb, T.; Montfort, F.; Kühn, J.; Aspert, N.; Cuche, E.; Marian, A.; Charrière, F.; Bourquin, S.; Marquet, P.; Depeursinge, C. Numerical parametric lens for shifting, magnification, and complete aberration compensation in digital holographic microscopy. J. Opt. Soc. Am. A 2006, 23, 3177–3190. [Google Scholar] [CrossRef]
- Cuche, E.; Marquet, P.; Depeursinge, C. Spatial filtering for zero-order and twin-image elimination in digital off-axis holography. Appl. Opt. 2000, 39, 4070–4075. [Google Scholar] [CrossRef]
- Anand, A.; Chhaniwal, V.K.; Patel, N.R.; Javidi, B. Automatic identification of malaria-infected RBC with digital holographic microscopy using correlation algorithms. IEEE Photonics J. 2012, 4, 1456–1464. [Google Scholar] [CrossRef]
- Moon, I.; Anand, A.; Cruz, M.; Javidi, B. Identification of Malaria-Infected Red Blood Cells Via Digital Shearing Interferometry and Statistical Inference. IEEE Photonics J. 2013, 5, 6900207. [Google Scholar] [CrossRef]
- Doblas, A.; Roche, E.; Ampudia-Blasco, F.J.; Martínez-Corral, M.; Saavedra, G.; Garcia-Sucerquia, J. Diabetes screening by telecentric digital holographic microscopy. J. Microsc. 2016, 261, 285–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avidi, B.A.J.; Arkman, A.D.A.M.M.; Awat, S.I.R.; Imothy, T.; Onnor, O.C.; Nand, A.R.U.N.A.; Ndemariam, B.I.A. Sickle cell disease diagnosis based on spatio- temporal cell dynamics analysis using 3D printed shearing digital holographic microscopy. Opt. Express 2018, 26, 13614–13627. [Google Scholar]
- Mugnano, M.; Memmolo, P.; Miccio, L.; Merola, F.; Bianco, V.; Bramanti, A.; Gambale, A.; Russo, R.; Andolfo, I.; Iolascon, A.; et al. Label-Free Optical Marker for Red-Blood-Cell Phenotyping of Inherited Anemias. Anal. Chem. 2018, 90, 7495–7501. [Google Scholar] [CrossRef] [PubMed]
- Isola, P.; Zhu, J.; Zhou, T.; Efros, A.A. Image-to-Image Translation with Conditional Adversarial Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5967–5976. [Google Scholar]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Li, C.; Wand, M. Precomputed Real-Time Texture Synthesis with Markovian Generative Adversarial Networks. In Proceedings of the Computer Vision—ECCV 2016, Amsterdam, The Netherlands, 8–16 October 2016; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 702–716. [Google Scholar]
- Khalid, M.; Baber, J.; Kasi, M.K.; Bakhtyar, M.; Devi, V.; Sheikh, N. Empirical Evaluation of Activation Functions in Deep Convolution Neural Network for Facial Expression Recognition. In Proceedings of the 2020 43rd International Conference on Telecommunications and Signal Processing (TSP), Milan, Italy, 7–9 July 2020; pp. 204–207. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lect. Notes Comput. Sci. 2015, 9351, 12–20. [Google Scholar]
- Brownlee, J. Generative Adversarial Networks with Python: Deep Learning Generative Models for Image Synthesis and Image Translation; Machine Learning Mastery: San Francisco, CA, USA, 2019. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- cGAN QPI-DHM. Available online: https://oirl.github.io/cGAN-Digital-Holographic-microscopy/ (accessed on 20 November 2021).
- Goldstein, R.M.; Werner, C.L. Satellite radar interferometry Two-dimensional phase unwrapping. Radio Sci. 1988, 23, 713–720. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Karlsson, A.; Swartling, J.; Andersson-Engels, S. Light scattering by multiple red blood cells. J. Opt. Soc. Am. A 2004, 21, 1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trujillo, C.; Doblas, A.; Saavedra, G.; Martínez-Corral, M.; García-Sucerquia, J. Phase-shifting by means of an electronically tunable lens: Quantitative phase imaging of biological specimens with digital holographic microscopy. Opt. Lett. 2016, 41, 1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes-Rounds, C.; Bogue-Jimenez, B.; Garcia-Sucerquia, J.I.; Skalli, O.; Doblas, A. Advantages of Fresnel biprism-based digital holographic microscopy in quantitative phase imaging. J. Biomed. Opt. 2020, 25, 1–11. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castaneda, R.; Trujillo, C.; Doblas, A. Video-Rate Quantitative Phase Imaging Using a Digital Holographic Microscope and a Generative Adversarial Network. Sensors 2021, 21, 8021. https://doi.org/10.3390/s21238021
Castaneda R, Trujillo C, Doblas A. Video-Rate Quantitative Phase Imaging Using a Digital Holographic Microscope and a Generative Adversarial Network. Sensors. 2021; 21(23):8021. https://doi.org/10.3390/s21238021
Chicago/Turabian StyleCastaneda, Raul, Carlos Trujillo, and Ana Doblas. 2021. "Video-Rate Quantitative Phase Imaging Using a Digital Holographic Microscope and a Generative Adversarial Network" Sensors 21, no. 23: 8021. https://doi.org/10.3390/s21238021
APA StyleCastaneda, R., Trujillo, C., & Doblas, A. (2021). Video-Rate Quantitative Phase Imaging Using a Digital Holographic Microscope and a Generative Adversarial Network. Sensors, 21(23), 8021. https://doi.org/10.3390/s21238021







