Machine Learning-Based Plant Detection Algorithms to Automate Counting Tasks Using 3D Canopy Scans
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Cultivation Conditions, Ground Truth Measurements
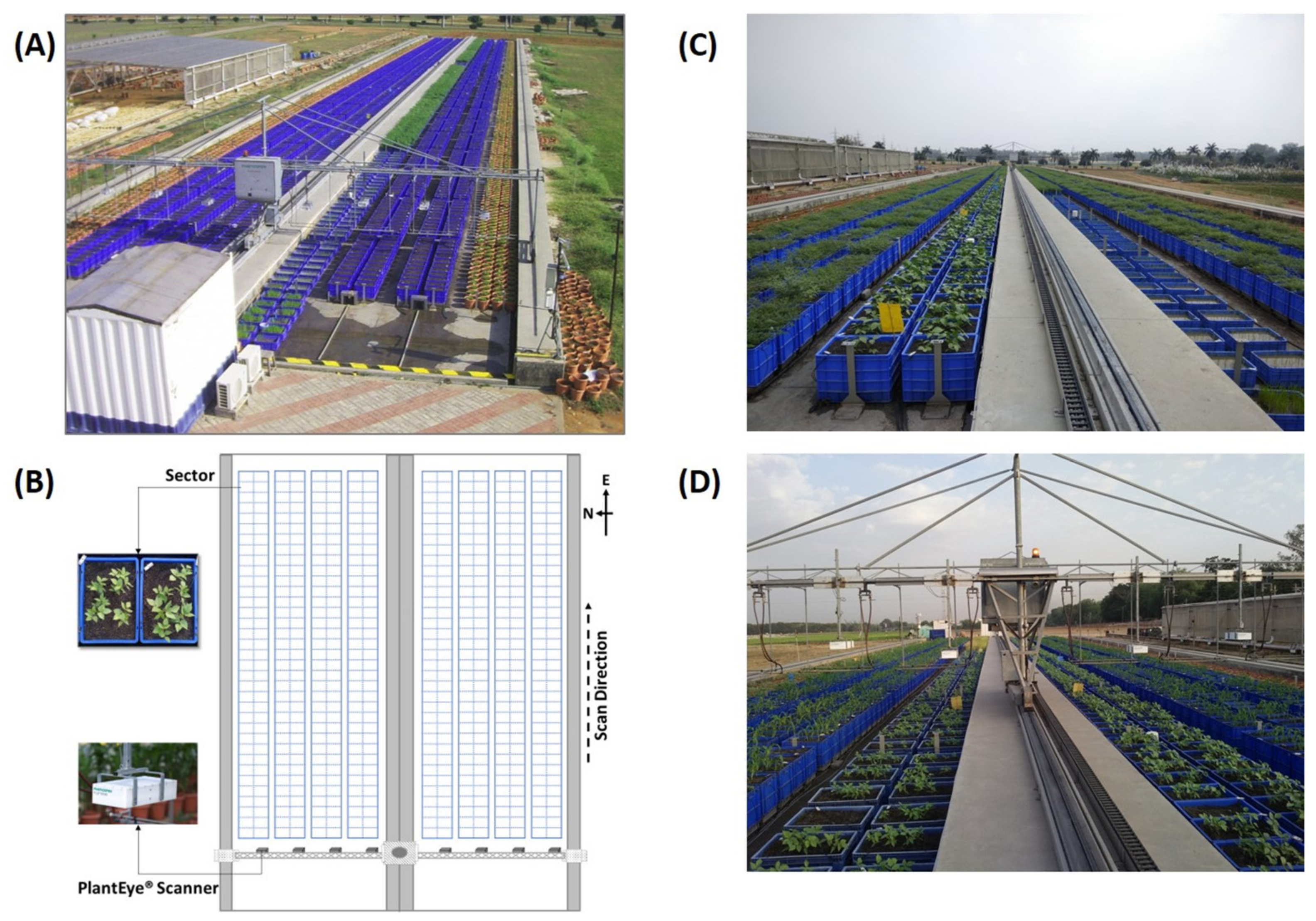
2.2. Basic Setup and Scanning Protocol
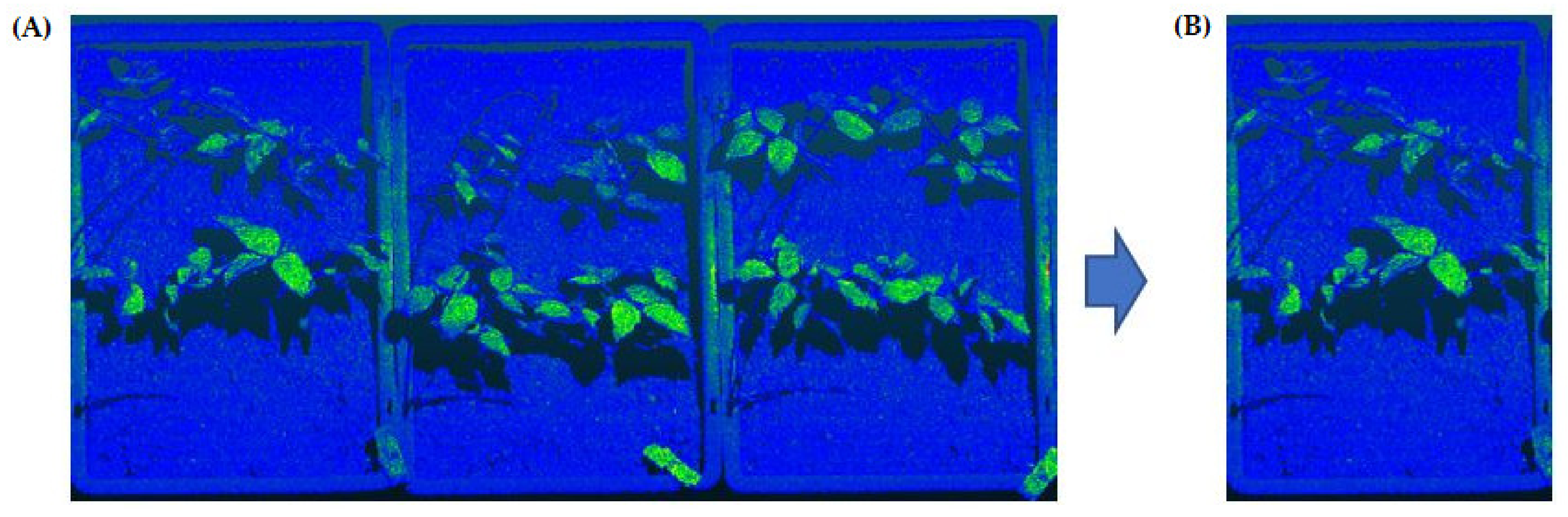
2.3. Tray Segmentation (Step 1)
2.4. Soil Segmentation (Step 2)
2.4.1. Region Growing Segmentation (RGS)
2.4.2. Random Sample Consensus (RANSAC) Model
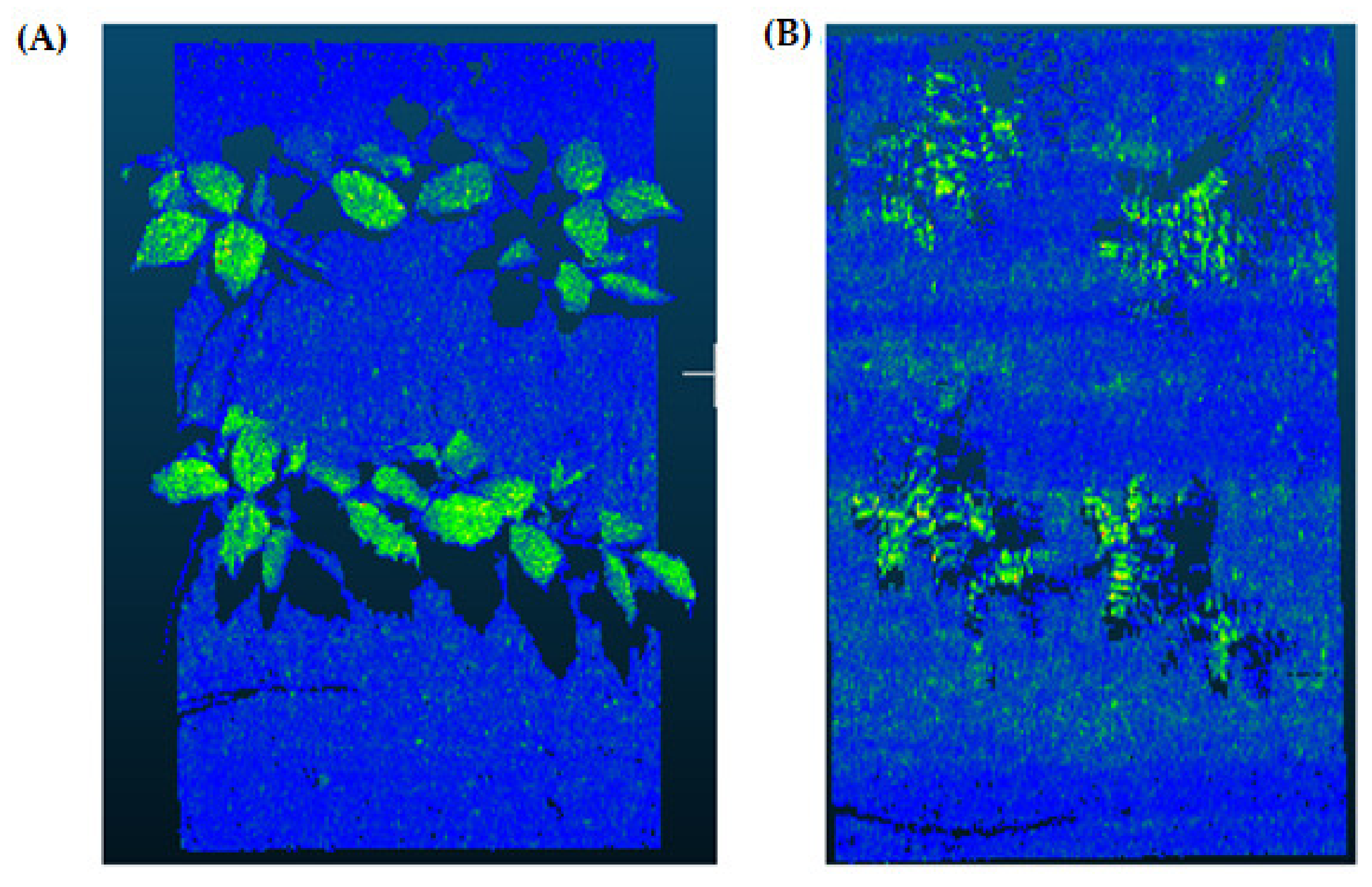
2.5. Rasterization (Step 3)
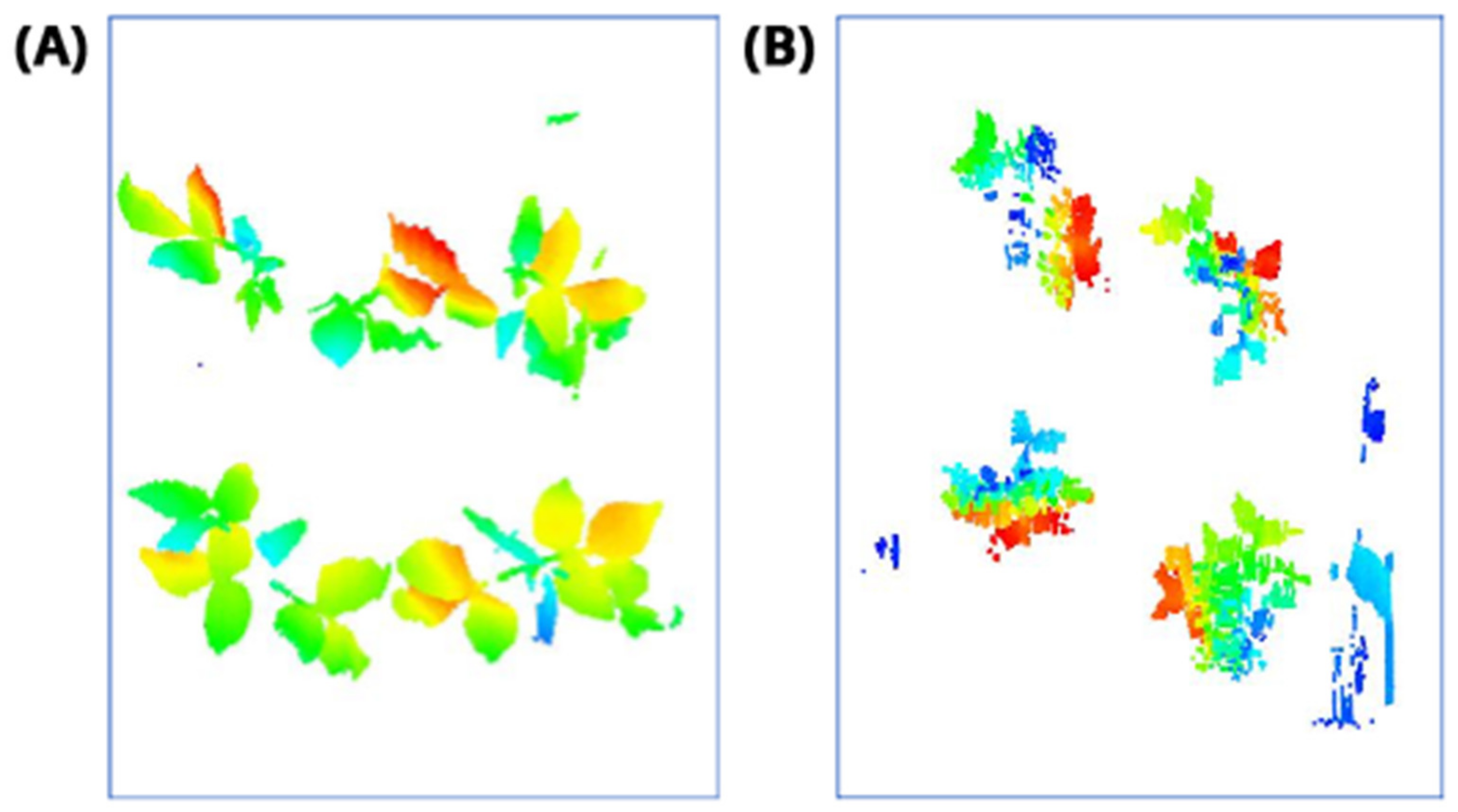
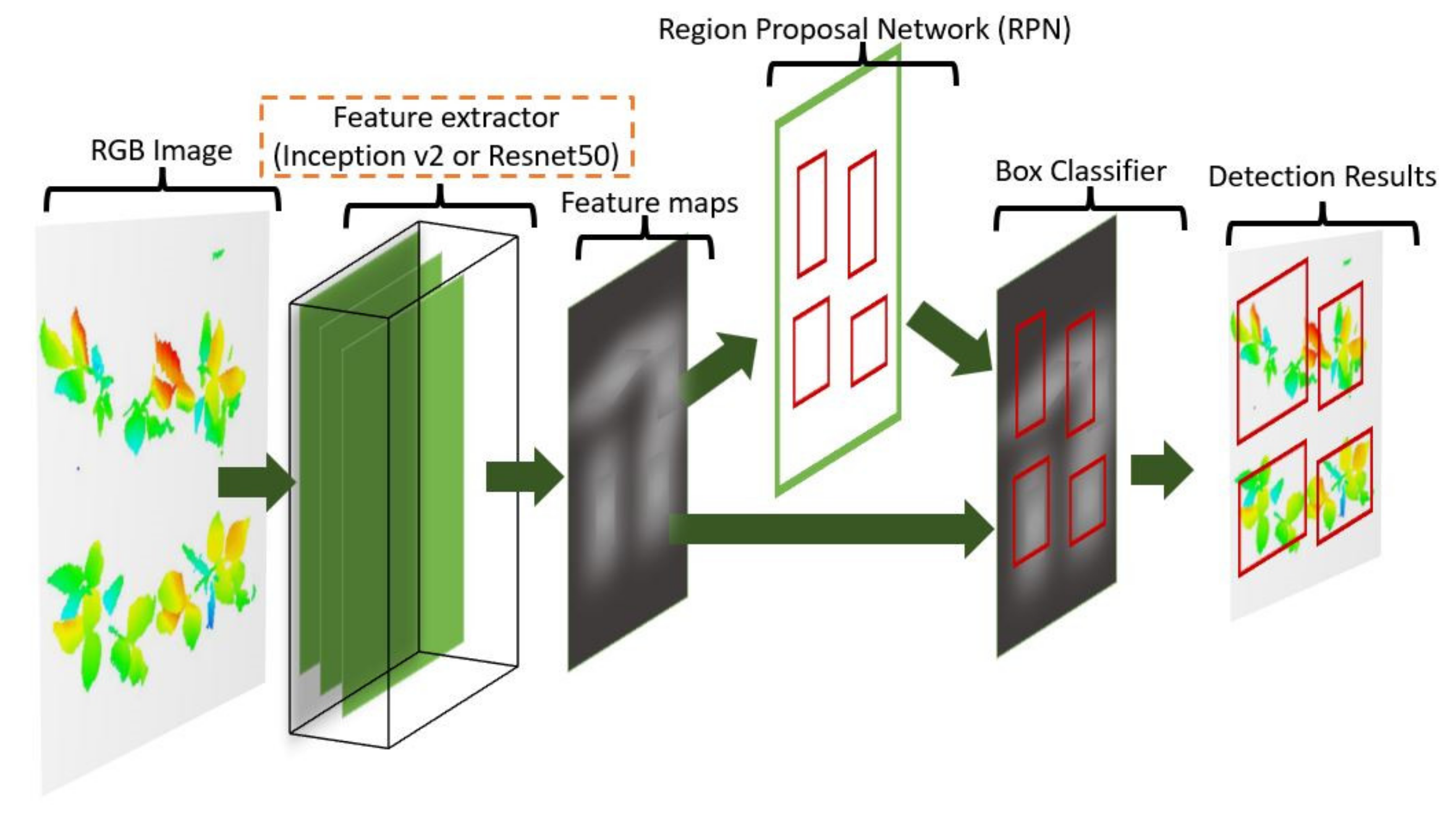
2.6. Plant Detection and Counting (Steps 4–5)
2.6.1. Convolutional Neural Networks (CNNs) for Object Detection
2.6.2. Image Labeling and Dataset Production
2.6.3. Training and Models Configuration
2.7. Performance Evaluation
3. Results
4. Discussion
4.1. Overview
4.2. Use of 3D Point Clouds for Data Preprocessing
4.3. Implemented CNN Models for Data with Reduced Dimensionality to Recognize Individual Plants and Count Them
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tardieu, F.; Cabrera-Bosquet, L.; Pridmore, T.; Bennett, M. Plant phenomics, from sensors to knowledge. Curr. Biol. 2017, 27, R770–R783. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A Review of Imaging Techniques for Plant Phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Pommier, C.; Garnett, T.; Lawrence-Dill, C.J.; Pridmore, T.; Watt, M.; Pieruschka, R.; Ghamkhar, K. Editorial: Phenotyping; From Plant, to Data, to Impact and Highlights of the International Plant Phenotyping Symposium-IPPS 2018. Front. Plant Sci. 2020, 11, 1907. [Google Scholar] [CrossRef] [PubMed]
- Kholová, J.; Urban, M.O.; Cock, J.; Arcos, J.; Arnaud, E.; Aytekin, D.; Azevedo, V.; Barnes, A.P.; Ceccarelli, S.; Chavarriaga, P.; et al. In pursuit of a better world: Crop improvement and the CGIAR. J. Exp. Bot. 2021, 72, 5158–5179. [Google Scholar] [CrossRef] [PubMed]
- Vadez, V.; Kholová, J.; Hummel, G.; Zhokhavets, U.; Gupta, S.K.; Hash, C.T. LeasyScan: A novel concept combining 3D imaging and lysimetry for high-throughput phenotyping of traits controlling plant water budget. J. Exp. Bot. 2015, 66, 5581–5593. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics—Technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Brown, T.B.; Cheng, R.; Sirault, X.R.; Rungrat, T.; Murray, K.D.; Trtilek, M.; Furbank, R.T.; Badger, M.; Pogson, B.J.; Borevitz, J.O. TraitCapture: Genomic and environment modelling of plant phenomic data. Curr. Opin. Plant. Biol. 2014, 18, 73–79. [Google Scholar] [CrossRef]
- Virlet, N.; Sabermanesh, K.; Sadeghi-Tehran, P.; Hawkesford, M.J. Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 2016, 44, 143–153. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future Scenarios for Plant Phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef]
- Tardieu, F.; Hammer, G. Designing crops for new challenges. Eur. J. Agron. 2012, 42, 1–2. [Google Scholar] [CrossRef]
- Tardieu, F.; Simonneau, T.; Muller, B. The Physiological Basis of Drought Tolerance in Crop Plants: A Scenario-Dependent Probabilistic Approach. Annu. Rev. Plant Biol. 2018, 69, 733–759. [Google Scholar] [CrossRef]
- Kholová, J.; Murugesan, T.; Kaliamoorthy, S.; Malayee, S.; Baddam, R.; Hammer, G.L.; McLean, G.; Deshpande, S.; Hash, C.T.; Craufurd, P.Q.; et al. Modelling the effect of plant water use traits on yield and stay-green expression in sorghum. Funct. Plant Biol. 2014, 41, 1019–1034. [Google Scholar] [CrossRef]
- Sivasakthi, K.; Thudi, M.; Tharanya, M.; Kale, S.M.; Kholová, J.; Halime, M.H.; Jaganathan, D.; Baddam, R.; Thirunalasundari, T.; Gaur, P.M.; et al. Plant vigour QTLs co-map with an earlier reported QTL hotspot for drought tolerance while water saving QTLs map in other regions of the chickpea genome. BMC Plant Biol. 2018, 18, 29. [Google Scholar] [CrossRef]
- Sivasakthi, K.; Marques, E.; Kalungwana, N.; Carrasquilla-Garcia, N.; Chang, P.L.; Bergmann, E.M.; Bueno, E.; Cordeiro, M.; Sani, S.G.A.; Udupa, S.M.; et al. Functional Dissection of the Chickpea (Cicer arietinum L.) Stay-Green Phenotype Associated with Molecular Variation at an Ortholog of Mendel’s I Gene for Cotyledon Color: Implications for Crop Production and Carotenoid Biofortification. Int. J. Mol. Sci. 2019, 20, 5562. [Google Scholar] [CrossRef]
- Tharanya, M.; Kholova, J.; Sivasakthi, K.; Seghal, D.; Hash, C.T.; Raj, B.; Srivastava, R.K.; Baddam, R.; Thirunalasundari, T.; Yadav, R.; et al. Quantitative trait loci (QTLs) for water use and crop production traits co-locate with major QTL for tolerance to water deficit in a fine-mapping population of pearl millet (Pennisetum glaucum L. R.Br.). Theor. Appl. Genet. 2018, 131, 1509–1529. [Google Scholar] [CrossRef] [PubMed]
- Kar, S.; Garin, V.; Kholová, J.; Vadez, V.; Durbha, S.S.; Tanaka, R.; Iwata, H.; Urban, M.O.; Adinarayana, J. SpaTemHTP: A Data Analysis Pipeline for Efficient Processing and Utilization of Temporal High-Throughput Phenotyping Data. Front. Plant Sci. 2020, 11, 552509. [Google Scholar] [CrossRef]
- Kar, S.; Tanaka, R.; Korbu, L.B.; Kholová, J.; Iwata, H.; Durbha, S.S.; Adinarayana, J.; Vadez, V. Automated discretization of ‘transpiration restriction to increasing VPD’ features from outdoors high-throughput phenotyping data. Plant Methods 2020, 16, 140. [Google Scholar] [CrossRef]
- Fanourakis, D.; Briese, C.; Max, J.F.; Kleinen, S.; Putz, A.; Fiorani, F.; Ulbrich, A.; Schurr, U. Rapid determination of leaf area and plant height by using light curtain arrays in four species with contrasting shoot architecture. Plant Methods 2014, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Pound, M.P.; Atkinson, J.; Townsend, A.J.; Wilson, M.; Griffiths, M.; Jackson, A.; Bulat, A.; Tzimiropoulos, G.; Wells, D.; Murchie, E.; et al. Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. GigaScience 2017, 6, gix083. [Google Scholar] [CrossRef] [PubMed]
- Grinblat, G.L.; Uzal, L.C.; Larese, M.G.; Granitto, P. Deep learning for plant identification using vein morphological patterns. Comput. Electron. Agric. 2016, 127, 418–424. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.; Wang, G.; Zhang, H. Deep Learning for Plant Identification in Natural Environment. Comput. Intell. Neurosci. 2017, 2017, 7361042. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, J.; Pajares, G.; Montalvo, M.; Romeo, J.; Guijarro, M. Support Vector Machines for crop/weeds identification in maize fields. Expert Syst. Appl. 2012, 39, 11149–11155. [Google Scholar] [CrossRef]
- Tellaeche, A.; Burgos-Artizzu, X.P.; Pajares, G.; Ribeiro, A. A vision-based method for weeds identification through the Bayesian decision theory. Pattern Recognit. 2008, 41, 521–530. [Google Scholar] [CrossRef]
- Sakamoto, T.; Gitelson, A.A.; Nguy-Robertson, A.L.; Arkebauer, T.J.; Wardlow, B.D.; Suyker, A.E.; Verma, S.B.; Shibayama, M. An alternative method using digital cameras for continuous monitoring of crop status. Agric. For. Meteorol. 2012, 154, 113–126. [Google Scholar] [CrossRef]
- Vega, F.A.; Carvajal-Ramírez, F.; Pérez-Saiz, M.; Rosúa, F.O. Multi-temporal imaging using an unmanned aerial vehicle for monitoring a sunflower crop. Biosyst. Eng. 2015, 132, 19–27. [Google Scholar] [CrossRef]
- Yeh, Y.-H.F.; Lai, T.-C.; Liu, T.-Y.; Liu, C.-C.; Chung, W.-C.; Lin, T.-T. An automated growth measurement system for leafy vegetables. Biosyst. Eng. 2014, 117, 43–50. [Google Scholar] [CrossRef]
- Gong, A.; Yu, J.; He, Y.; Qiu, Z. Citrus yield estimation based on images processed by an Android mobile phone. Biosyst. Eng. 2013, 115, 162–170. [Google Scholar] [CrossRef]
- Payne, A.; Walsh, K.; Subedi, P.; Jarvis, D. Estimation of mango crop yield using image analysis—Segmentation method. Comput. Electron. Agric. 2013, 91, 57–64. [Google Scholar] [CrossRef]
- Polder, G.; van der Heijden, G.W.; van Doorn, J.; Baltissen, T.A. Automatic detection of tulip breaking virus (TBV) in tulip fields using machine vision. Biosyst. Eng. 2014, 117, 35–42. [Google Scholar] [CrossRef]
- Pourreza, A.; Lee, W.S.; Etxeberria, E.; Banerjee, A. An evaluation of a vision-based sensor performance in Huanglongbing disease identification. Biosyst. Eng. 2015, 130, 13–22. [Google Scholar] [CrossRef]
- Valiente-González, J.M.; Andreu-García, G.; Potter, P.; Rodas-Jordá, A. Automatic corn (Zea mays) kernel inspection system using novelty detection based on principal component analysis. Biosyst. Eng. 2014, 117, 94–103. [Google Scholar] [CrossRef]
- Pavlíček, J.; Jarolímek, J.; Jarolímek, J.; Pavlíčková, P.; Dvořák, S.; Pavlík, J.; Hanzlík, P. Automated Wildlife Recognition. Agris-Line Pap. Econ. Inform. 2018, 10, 51–60. [Google Scholar] [CrossRef][Green Version]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Itakura, K.; Hosoi, F. Automatic individual tree detection and canopy segmentation from three-dimensional point cloud images obtained from ground-based lidar. J. Agric. Meteorol. 2018, 74, 109–113. [Google Scholar] [CrossRef]
- Malambo, L.; Popescu, S.; Horne, D.; Pugh, N.; Rooney, W. Automated detection and measurement of individual sorghum panicles using density-based clustering of terrestrial lidar data. ISPRS J. Photogramm. Remote Sens. 2019, 149, 1–13. [Google Scholar] [CrossRef]
- Weiss, U.; Biber, P. Plant detection and mapping for agricultural robots using a 3D LIDAR sensor. Robot. Auton. Syst. 2011, 59, 265–273. [Google Scholar] [CrossRef]
- Ugarriza, L.G.; Saber, E.; Vantaram, S.R.; Amuso, V.; Shaw, M.; Bhaskar, R. Automatic Image Segmentation by Dynamic Region Growth and Multiresolution Merging. IEEE Trans. Image Process. 2009, 18, 2275–2288. [Google Scholar] [CrossRef]
- Zeineldin, R.A.; El-Fishawy, N.A. A Survey of RANSAC enhancements for Plane Detection in 3D Point Clouds. Menoufia J. Electron. Eng. Res. 2017, 26, 519–537. [Google Scholar] [CrossRef]
- Rusu, R.B. Semantic 3D Object Maps for Everyday Manipulation in Human Living Environments. KI Künstliche Intell. 2010, 24, 345–348. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Maturana, D.; Scherer, S. VoxNet: VoxNet: A 3D convolutional neural network for real-time object recognition. In Proceedings of the IEEE International Conference on Intelligent Robots and Systems, Hamburg, Germany, 25 September–2 October 2015. [Google Scholar]
- Kim, C.; Lee, J.; Han, T.; Kim, Y.-M. A hybrid framework combining background subtraction and deep neural networks for rapid person detection. J. Big Data 2018, 5, 22. [Google Scholar] [CrossRef]
- Mohamed, S.S.; Tahir, N.M.; Adnan, R. Background modelling and background subtraction performance for object detection. In Proceedings of the 2010 6th International Colloquium on Signal Processing and Its Applications (CSPA 2010), Malacca, Malaysia, 21–23 May 2010. [Google Scholar]
- Chen, S.; Zheng, L.; Zhang, Y.; Sun, Z.; Xu, K. VERAM: Rapid determination of leaf area and plant height by using light curtain arrays in four species with contrasting shoot architecture. IEEE Trans. Vis. Comput. Graph. 2019, 25, 3244–3257. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. Pointnet: Deep learning on point sets for 3D classification and segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Yavartanoo, M.; Kim, E.Y.; Lee, K.M. SPNet: Deep 3D object classification and retrieval using stereographic projection. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Xie, Y.; Tian, J.; Zhu, X.X. Linking Points with Labels in 3D: A Review of Point Cloud Semantic Segmentation. IEEE Geosci. Remote Sens. Mag. 2020, 8, 38–59. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on Machine Learning (ICML), Lille, France, 7–9 July 2015. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- He, K.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]






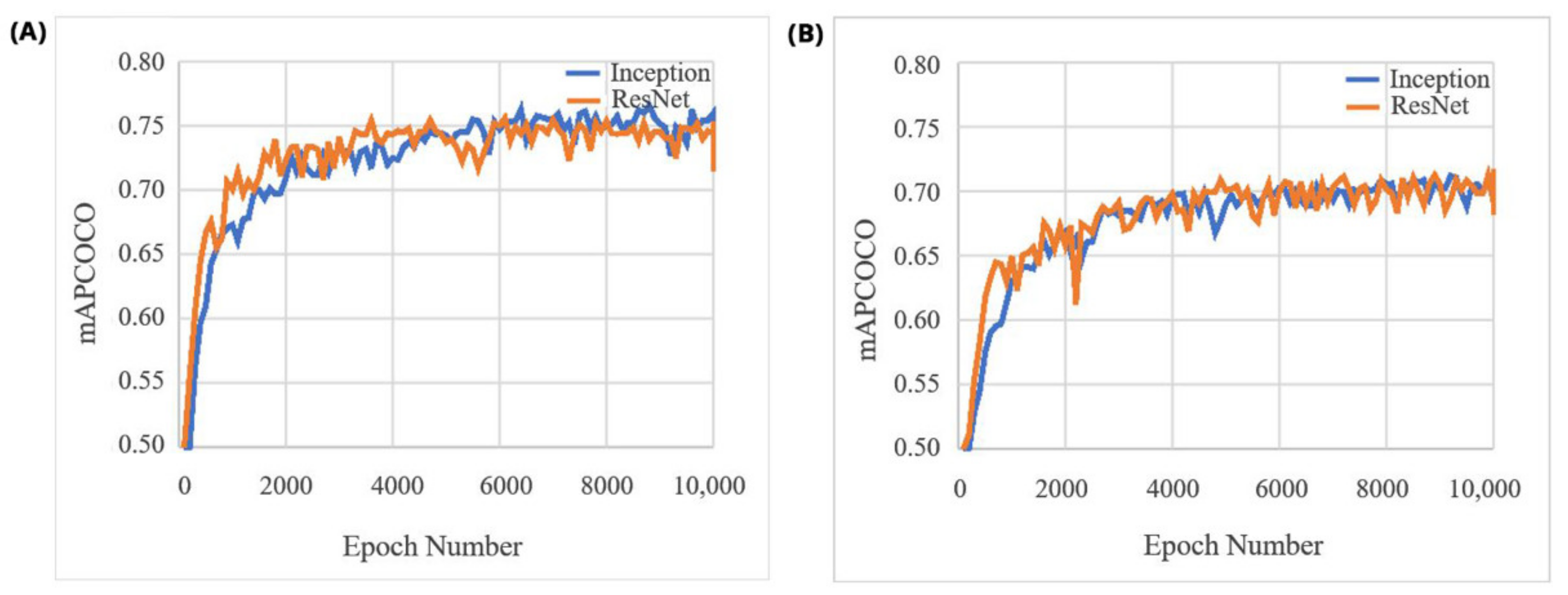
| Algorithm | COCO mAPCOCO | Bean mAPCOCO | Chickpea mAPCOCO |
|---|---|---|---|
| Faster RCNN Inception-v2 | 0.28 | 0.7525 | 0.7108 |
| Faster RCNN ResNet50 | 0.30 | 0.7477 | 0.7075 |
| Test Images (Tray Count) | Plant Count | MAPE | ||||
|---|---|---|---|---|---|---|
| Manual Counting | Inception-v2 Prediction | ResNet50 Prediction | Inception-v2 | ResNet50 | ||
| Mung bean | 72 | 275 | 287 | 303 | 6.82 | 11.80 |
| Chickpea | 237 | 1606 | 1547 | 1620 | 8.17 | 7.13 |
| Inception-v2 | ResNet50 | Ground-Truth | |
|---|---|---|---|
| 1 | 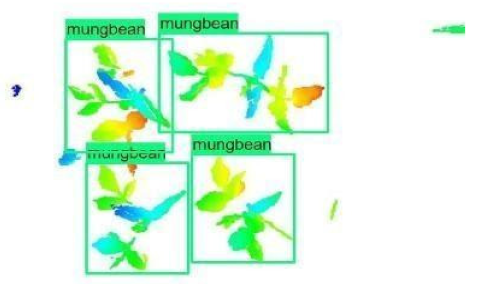 | 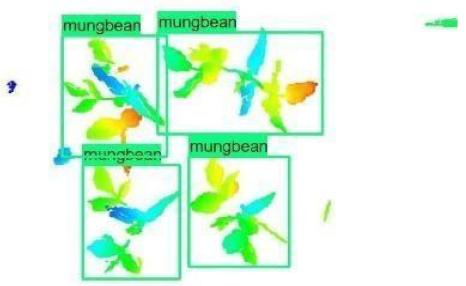 |  |
| 2 | 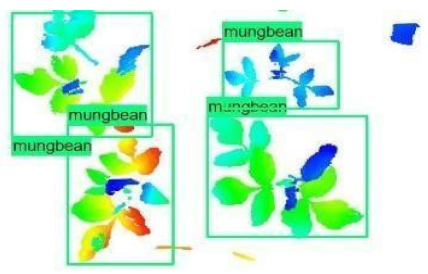 | 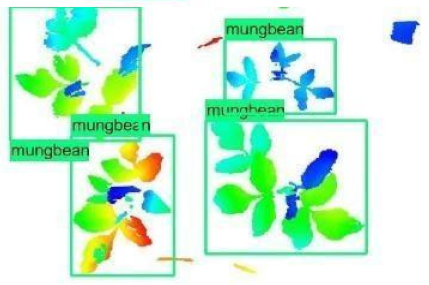 |  |
| 3 |  | 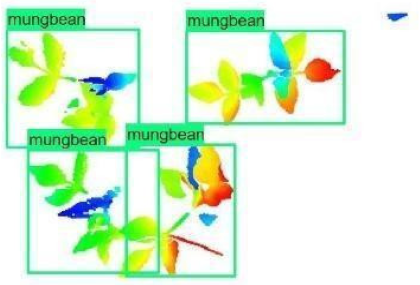 |  |
| 4 |  | 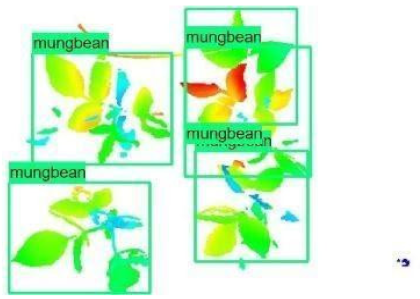 |  |
| 5 |  |  |  |
| 6 |  |  |  |
| 7 | 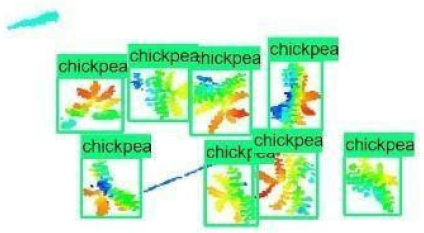 | 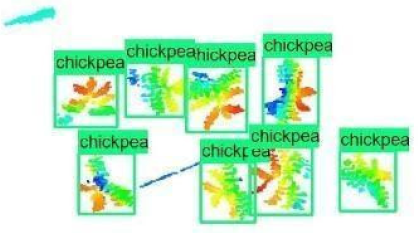 |  |
| 8 |  | 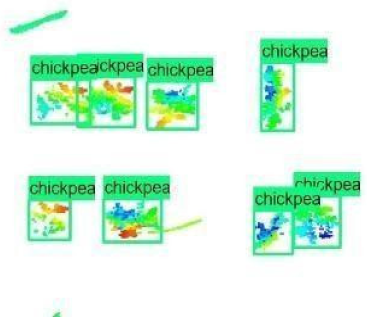 |  |
| Training Time | Plant Counting Time | |
|---|---|---|
| Faster RCNN Inception-v2 | 1.25 h | 15 s |
| Faster RCNN ResNet50 | 1.45 h | 18 s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kartal, S.; Choudhary, S.; Masner, J.; Kholová, J.; Stočes, M.; Gattu, P.; Schwartz, S.; Kissel, E. Machine Learning-Based Plant Detection Algorithms to Automate Counting Tasks Using 3D Canopy Scans. Sensors 2021, 21, 8022. https://doi.org/10.3390/s21238022
Kartal S, Choudhary S, Masner J, Kholová J, Stočes M, Gattu P, Schwartz S, Kissel E. Machine Learning-Based Plant Detection Algorithms to Automate Counting Tasks Using 3D Canopy Scans. Sensors. 2021; 21(23):8022. https://doi.org/10.3390/s21238022
Chicago/Turabian StyleKartal, Serkan, Sunita Choudhary, Jan Masner, Jana Kholová, Michal Stočes, Priyanka Gattu, Stefan Schwartz, and Ewaut Kissel. 2021. "Machine Learning-Based Plant Detection Algorithms to Automate Counting Tasks Using 3D Canopy Scans" Sensors 21, no. 23: 8022. https://doi.org/10.3390/s21238022
APA StyleKartal, S., Choudhary, S., Masner, J., Kholová, J., Stočes, M., Gattu, P., Schwartz, S., & Kissel, E. (2021). Machine Learning-Based Plant Detection Algorithms to Automate Counting Tasks Using 3D Canopy Scans. Sensors, 21(23), 8022. https://doi.org/10.3390/s21238022






