Graphene-Based Sensor for Detection of Bacterial Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chip Fabrication
2.2. Culture Media and Bacterial Growth
2.3. Set-Up and Evaluation of Bacterial Sensing Ability
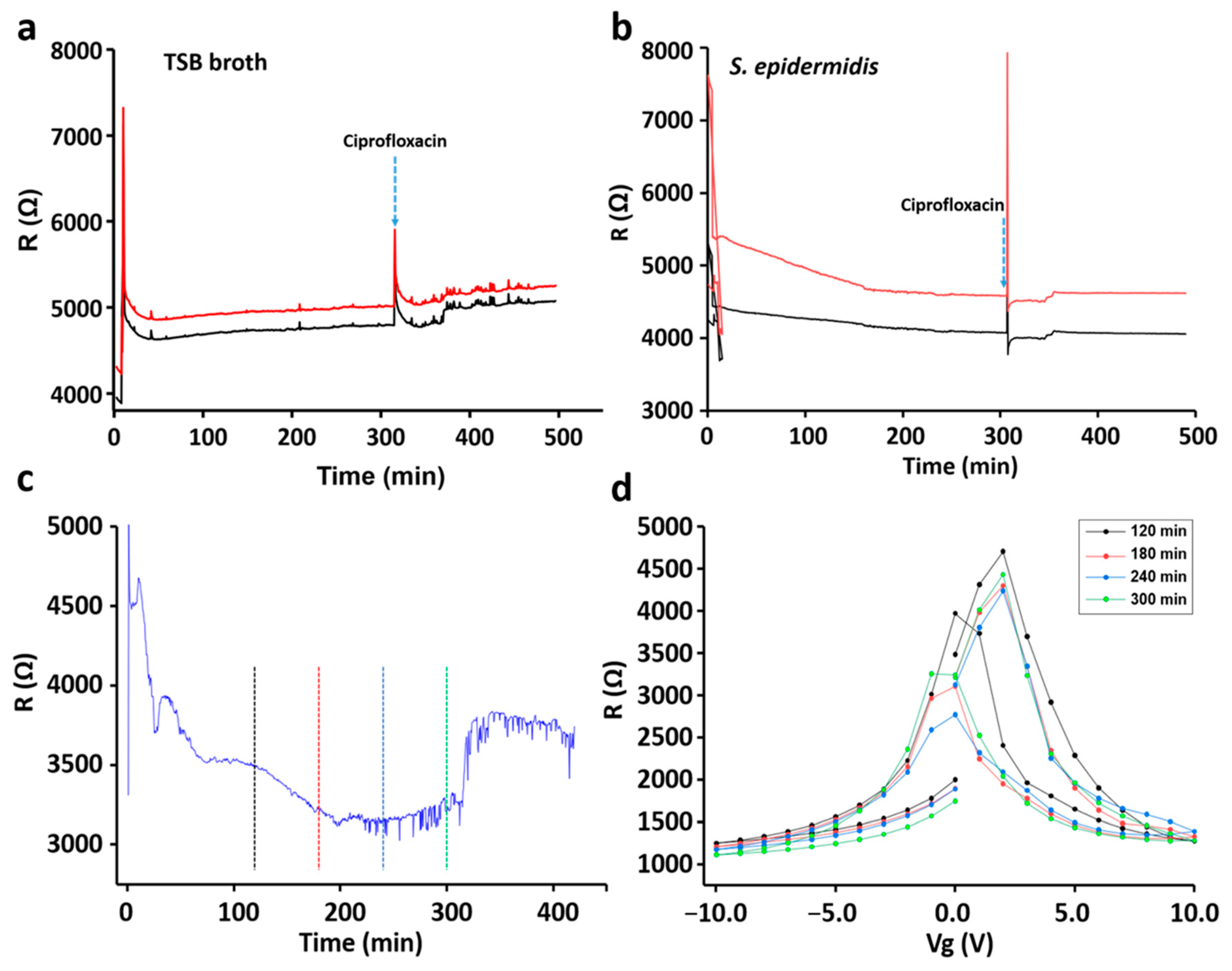
3. Results
3.1. Characterization and Validation of the Sensor Chip
3.2. Bacterial Growth Pattern
3.3. Sensing of Different Bacterial Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fonkwo, P.N. Pricing infectious disease. The economic and health implications of infectious diseases. EMBO Rep. 2008, 9 (Suppl. 1), S13–S17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Haque, M.; Sartelli, M.; McKimm, J.; Bakar, M.A. Health care-associated infections—An overview. Infect. Drug. Resist. 2018, 11, 2321–2333. [Google Scholar] [CrossRef] [Green Version]
- Atrazhev, A.; Manage, D.P.; Stickel, A.J.; Crabtree, H.J.; Pilarski, L.M.; Acker, J.P. In-Gel technology for PCR genotyping and pathogen detection. Anal. Chem. 2010, 82, 8079–8087. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Rothberg, L.J. DNA sequence detection using selective fluorescence quenching of tagged oligonucleotide probes by gold nanoparticles. Anal. Chem. 2004, 76, 5414–5417. [Google Scholar] [CrossRef]
- Charlermroj, R.; Himananto, O.; Seepiban, C.; Kumpoosiri, M.; Warin, N.; Oplatowska, M.; Gajanandana, O.; Grant, I.R.; Karoonuthaisiri, N.; Elliott, C.T. Multiplex detection of plant pathogens using a microsphere immunoassay technology. PLoS ONE 2013, 8, e62344. [Google Scholar] [CrossRef] [Green Version]
- Peham, J.R.; Recnik, L.M.; Grienauer, W.; Vellekoop, M.J.; Nohammer, C.; Wiesinger-Mayr, H. Disposable microfluidic chip for rapid pathogen identification with DNA microarrays. Micro. Technol. 2012, 18, 311–318. [Google Scholar] [CrossRef]
- Vimont, A.; Vernozy-Rozand, C.; Montet, M.P.; Lazizzera, C.; Bavai, C.; Delignette-Muller, M.L. Modeling and predicting the simultaneous growth of Escherichia coli O157:H7 and ground beef background microflora for various enrichment protocols. Appl. Environ. Microbiol. 2006, 72, 261–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruno, J.; Carrillo, M.; Phillips, T.; Andrews, C. A novel screening method for competitive FRET-aptamers applied to E. coli assay development. J. Fluor. 2010, 20, 1211–1223. [Google Scholar] [CrossRef] [PubMed]
- Ohk, S.H.; Koo, O.K.; Sen, T.; Yamamoto, C.M.; Bhunia, A.K. Antibody-aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food. J. Appl. Microbiol. 2010, 109, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Torres-Chavolla, E.; Alocilja, E.C. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron. 2009, 24, 3175–3182. [Google Scholar] [CrossRef]
- Li, Y.; Luo, G.; Qing, Z.; Li, X.; Zou, Z.; Yang, R. Colorimetric aminotriazole assay based on catalase deactivation-dependent longitudinal etching of gold nanorods. Microchim. Acta 2019, 186, 565. [Google Scholar] [CrossRef]
- Qing, Z.; Bai, A.; Chen, L.; Xing, S.; Zou, Z.; Lei, Y.; Li, J.; Liu, J.; Yang, R. An activatable nanoenzyme reactor for coenhanced chemodynamic and starving therapy against tumor hypoxia and antioxidant defense system. CCS Chem. 2021, 3, 1217–1230. [Google Scholar] [CrossRef]
- Qing, Z.; Luo, G.; Xing, S.; Zou, Z.; Lei, Y.; Liu, J.; Yang, R. Pt–S bond-mediated nanoflares for high-fidelity intracellular applications by avoiding thiol cleavage. Angew. Chem. Int. Ed. 2020, 132, 14148–14152. [Google Scholar] [CrossRef]
- Sanvicens, N.; Pastells, C.; Pascual, N.; Marco, M.-P. Nanoparticle-based biosensors for detection of pathogenic bacteria. TrAC Trends Anal. Chem. 2009, 28, 1243–1252. [Google Scholar] [CrossRef]
- Justino, C.I.; Gomes, A.R.; Freitas, A.C.; Duarte, A.C.; Rocha Santos, T.A. Graphene based sensors and biosensors. TrAC Trends Anal. Chem. 2017, 91, 53–66. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H.; Aldalbahi, A.; Zuo, X.; Fan, C.; Mi, X. Fluorescent biosensors enabled by graphene and graphene oxide. Biosens. Bioelectron. 2017, 89, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Du, D.; Lin, Y. Graphene-like 2D nanomaterial-based biointerfaces for biosensing applications. Biosens. Bioelectron. 2017, 89, 43–55. [Google Scholar] [CrossRef]
- Forsyth, R.; Devadoss, A.; Guy, O.J. Graphene field effect transistors for biomedical applications: Current status and future prospects. Diagnostics 2017, 7, 45. [Google Scholar] [CrossRef] [Green Version]
- Kolev, S.K.; Aleksandrov, H.A.; Atanasov, V.A.; Popov, V.N.; Milenov, T.I. Interaction of graphene with out-of-plane aromatic hydrocarbons. J. Phys. Chem. C 2019, 123, 21448–21456. [Google Scholar] [CrossRef]
- Danielson, E.; Sontakke, V.A.; Porkovich, A.J.; Yokobayashi, Y.; Sowwan, M. Graphene based field-effect transistor biosensors functionalized using gas-phase synthesized gold nanoparticles. Sens. Actuators B Chem. 2020, 320, 128432. [Google Scholar] [CrossRef]
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for whole-cell bacterial detection. Clin. Microbiol. Rev. 2014, 27, 631–646. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Xie, X.; Duan, Y.; Wang, L.; Cheng, Z.; Cheng, J. A review of impedance measurements of whole cells. Biosens. Bioelectron. 2016, 77, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef]
- Guo, X.F.; Kulkarni, A.; Doepke, A.; Halsall, H.B.; Iyer, S.; Heineman, W.R. Carbohydrate-based label-free detection of Escherichia coli ORN 178 using electrochemical impedance spectroscopy. Anal. Chem. 2012, 84, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Lin, Z.; Zhang, D.; Wang, Y.; Hou, B. Impedimetric immunosensor doped with reduced graphene sheets fabricated by controllable electrodeposition for the non-labelled detection of bacteria. Biosens. Bioelectron. 2011, 26, 1959–1964. [Google Scholar] [CrossRef]
- Dweik, M.; Stringer, R.C.; Dastider, S.G.; Wu, Y.F.; Almasri, M.; Barizuddin, S. Specific and targeted detection of viable Escherichia coli O157:H7 using a sensitive and reusable impedance biosensor with dose and time response studies. Talanta 2012, 94, 84–89. [Google Scholar] [CrossRef]
- Wang, Y.X.; Ping, J.F.; Ye, Z.Z.; Wu, J.; Ying, Y.B. Impedimetric immunosensor based on gold nanoparticles modified graphene paper for label free detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2013, 49, 492–498. [Google Scholar] [CrossRef]
- Qi, P.; Wan, Y.; Zhang, D. Impedimetric biosensor based on cell mediated bioimprinted films for bacterial detection. Biosens. Bioelectron. 2013, 39, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Cremin, K.; Jones, B.A.; Teahan, J.; Meloni, G.N.; Perry, D.; Zerfass, C.; Asally, M.; Soyer, O.S.; Unwin, P.R. Scanning ion conductance microscopy reveals differences in the ionic environments of gram-positive and negative bacteria. Anal. Chem. 2020, 92, 16024–16032. [Google Scholar] [CrossRef]
- Pajerski, W.; Ochonska, D.; Brzychczy-Wloch, M.; Indyka, P.; Jarosz, M.; Gold-Cepa, M.; Sojka, Z.; Kotarba, A. Attachment efficiency of gold nanoparticles by Gram-positive and Gram-negative bacterial strains governed by surface charges. J. Nanopart. Res. 2019, 21, 186. [Google Scholar] [CrossRef] [Green Version]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.-E.; He, L.; Heo, J.; Hwang, G. Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front. Bioeng. Biotechnol. 2021, 9, 82. [Google Scholar] [CrossRef]
- De la Rosa, C.J.L.; Sun, J.; Lindvall, N.; Cole, M.T.; Nam, Y.; Loffler, M.; Olsson, E.; Teo, K.B.K.; Yurgens, A. Frame assisted H2O electrolysis induced H2 bubbling transfer of large area graphene grown by chemical vapor deposition on Cu. Appl. Phy. Lett. 2013, 102, 022101. [Google Scholar] [CrossRef] [Green Version]
- Vishnubhotla, R.; Ping, J.; Gao, Z.; Lee, A.; Saouaf, O.; Vrudhula, A.; Johnson, A.C. Scalable graphene aptasensors for drug quantification. AIP Adv. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Pandit, S.; Fazilati, M.; Gaska, K.; Derouiche, A.; Nypelö, T.; Mijakovic, I.; Kádár, R. The exo-polysaccharide component of extracellular matrix is essential for the viscoelastic properties of Bacillus subtilis biofilms. Int. J. Mol. Sci. 2020, 21, 6755. [Google Scholar] [CrossRef]
- Pandit, S.; Ravikumar, V.; Abdel-Haleem, A.M.; Derouiche, A.; Mokkapati, V.R.S.S.; Sihlbom, C.; Mineta, K.; Gojobori, T.; Gao, X.; Westerlund, F.; et al. Low concentrations of vitamin c reduce the synthesis of extracellular polymers and destabilize bacterial biofilms. Front. Microbiol. 2017, 8, 2599. [Google Scholar] [CrossRef] [Green Version]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef]
- Wu, G.; Meyyappan, M.; Lai, K.W.C. Simulation of graphene field-effect transistor biosensors for bacterial detection. Sensors 2018, 18, 1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, N.; Wang, W.; Ortiz-Marquez, J.C.; Catalano, M.; Gray, M.; Biglari, N.; Hikari, K.; Ling, X.; Gao, J.; van Opijnen, T.; et al. Dielectrophoresis assisted rapid, selective and single cell detection of antibiotic resistant bacteria with G-FETs. Biosens. Bioelectron. 2020, 156, 112123. [Google Scholar] [CrossRef] [PubMed]
- Sydow, A.; Krieg, T.; Mayer, F.; Schrader, J.; Holtmann, D. Electroactive bacteria—molecular mechanisms and genetic tools. Appl. Microbiol. Biotechnol. 2014, 98, 8481–8495. [Google Scholar] [CrossRef]
- Zhang, T.; Cui, C.; Chen, S.; Yang, H.; Shen, P. The direct electrocatalysis of Escherichia coli through electroactivated excretion in microbial fuel cell. Electrochem. Commun. 2008, 10, 293–297. [Google Scholar] [CrossRef]
- Rabaey, K.; Boon, N.; Höfte, M.; Verstraete, W. Microbial phenazine production enhances electron transfer in biofuel cells. Environ. Sci. Technol. 2005, 39, 3401–3408. [Google Scholar] [CrossRef]
- Barreiros, D.S.M.; Agusil, J.; Prieto-Simón, B.; Sporer, C.; Teixeira, V.; Samitier, J. Highly sensitive detection of pathogen Escherichia coli O157:H7 by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2013, 45, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Mohanta, G.C.; Deep, A. Bacteriophage immobilized graphene electrodes for impedimetric sensing of bacteria (Staphylococcus arlettae). Anal. Biochem. 2016, 505, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, H.; Vuong, C.; Vadyvaloo, V.; Wang, J.; Yao, Y.; Otto, M.; Gao, Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 2006, 74, 488–496. [Google Scholar] [CrossRef] [Green Version]
- De Kievit, T.R. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2009, 11, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Lee, K.-M.; Park, C.Y.; Kim, H.V.; Kim, Y.; Park, S. Quorum sensing is crucial to Escherichia coli O157:H7 biofilm formation under static or very slow laminar flow conditions. Biochip. J. 2016, 3, 241–250. [Google Scholar] [CrossRef]
- Gibson, B.; Wilson, D.J.; Feil, E.; Eyre-Walker, A. The distribution of bacterial doubling times in the wild. Proc. Biol. Sci. 2018, 285, 20180789. [Google Scholar] [CrossRef]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140–164. [Google Scholar] [CrossRef]
- Fey, P.D.; Olson, M.E. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010, 5, 917–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrova, O.E.; Sauer, K. Sticky situations: Key components that control bacterial surface attachment. J. Bacteriol. 2012, 194, 2413–2425. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Bacterial Strains | Culture pH (0 h) | Culture pH (5 h) |
|---|---|---|
| P. aeruginosa | 6.99 | 7.06 |
| S. epidermidis | 7.18 | 7.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandit, S.; Li, M.; Chen, Y.; Rahimi, S.; Mokkapati, V.; Merlo, A.; Yurgens, A.; Mijakovic, I. Graphene-Based Sensor for Detection of Bacterial Pathogens. Sensors 2021, 21, 8085. https://doi.org/10.3390/s21238085
Pandit S, Li M, Chen Y, Rahimi S, Mokkapati V, Merlo A, Yurgens A, Mijakovic I. Graphene-Based Sensor for Detection of Bacterial Pathogens. Sensors. 2021; 21(23):8085. https://doi.org/10.3390/s21238085
Chicago/Turabian StylePandit, Santosh, Mengyue Li, Yanyan Chen, Shadi Rahimi, Vrss Mokkapati, Alessandra Merlo, August Yurgens, and Ivan Mijakovic. 2021. "Graphene-Based Sensor for Detection of Bacterial Pathogens" Sensors 21, no. 23: 8085. https://doi.org/10.3390/s21238085
APA StylePandit, S., Li, M., Chen, Y., Rahimi, S., Mokkapati, V., Merlo, A., Yurgens, A., & Mijakovic, I. (2021). Graphene-Based Sensor for Detection of Bacterial Pathogens. Sensors, 21(23), 8085. https://doi.org/10.3390/s21238085







