DNA Electrochemical Biosensors for In Situ Probing of Pharmaceutical Drug Oxidative DNA Damage
Abstract
:1. Introduction
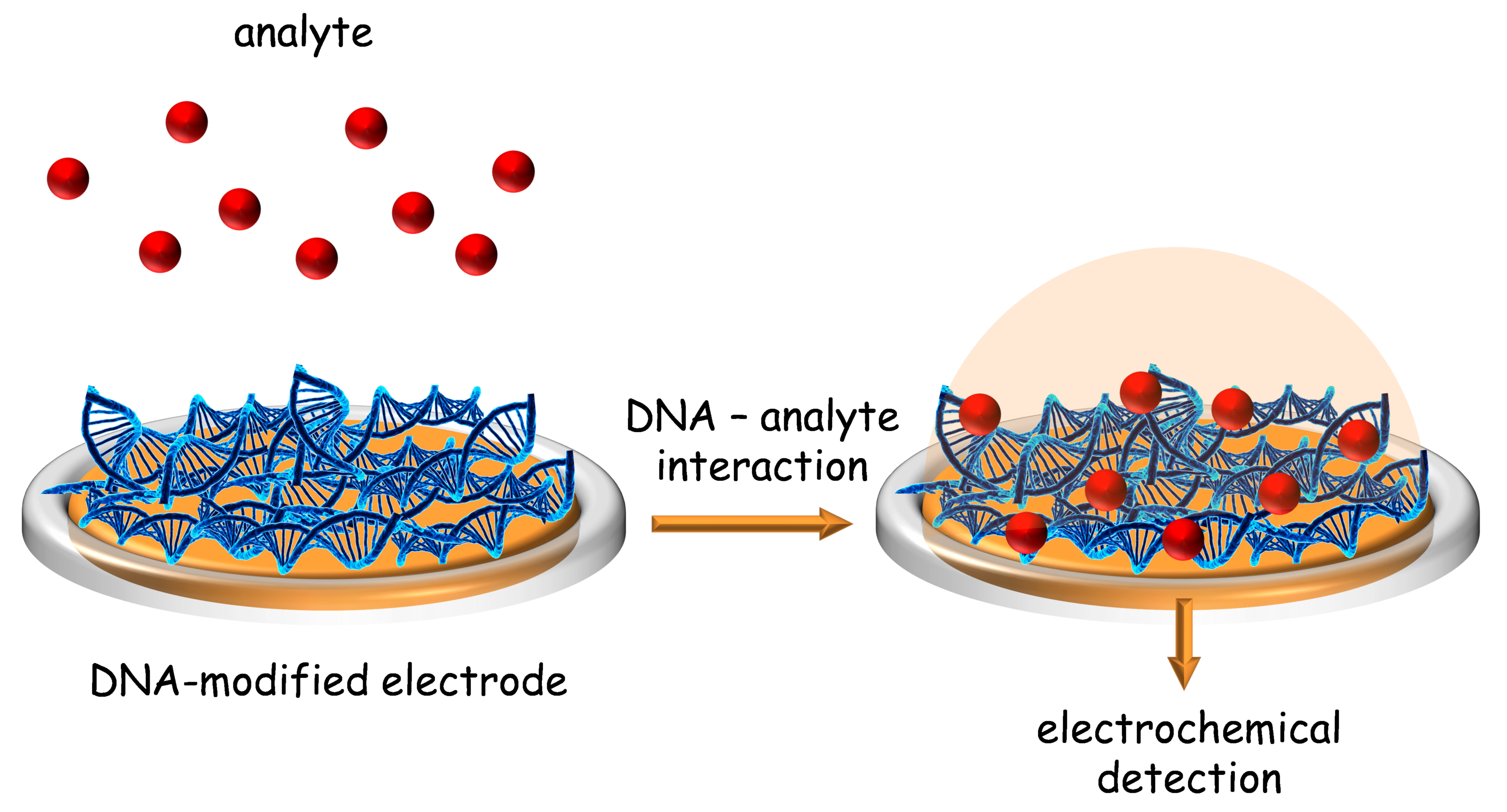
2. DNA Electrochemical Biosensor Characterization
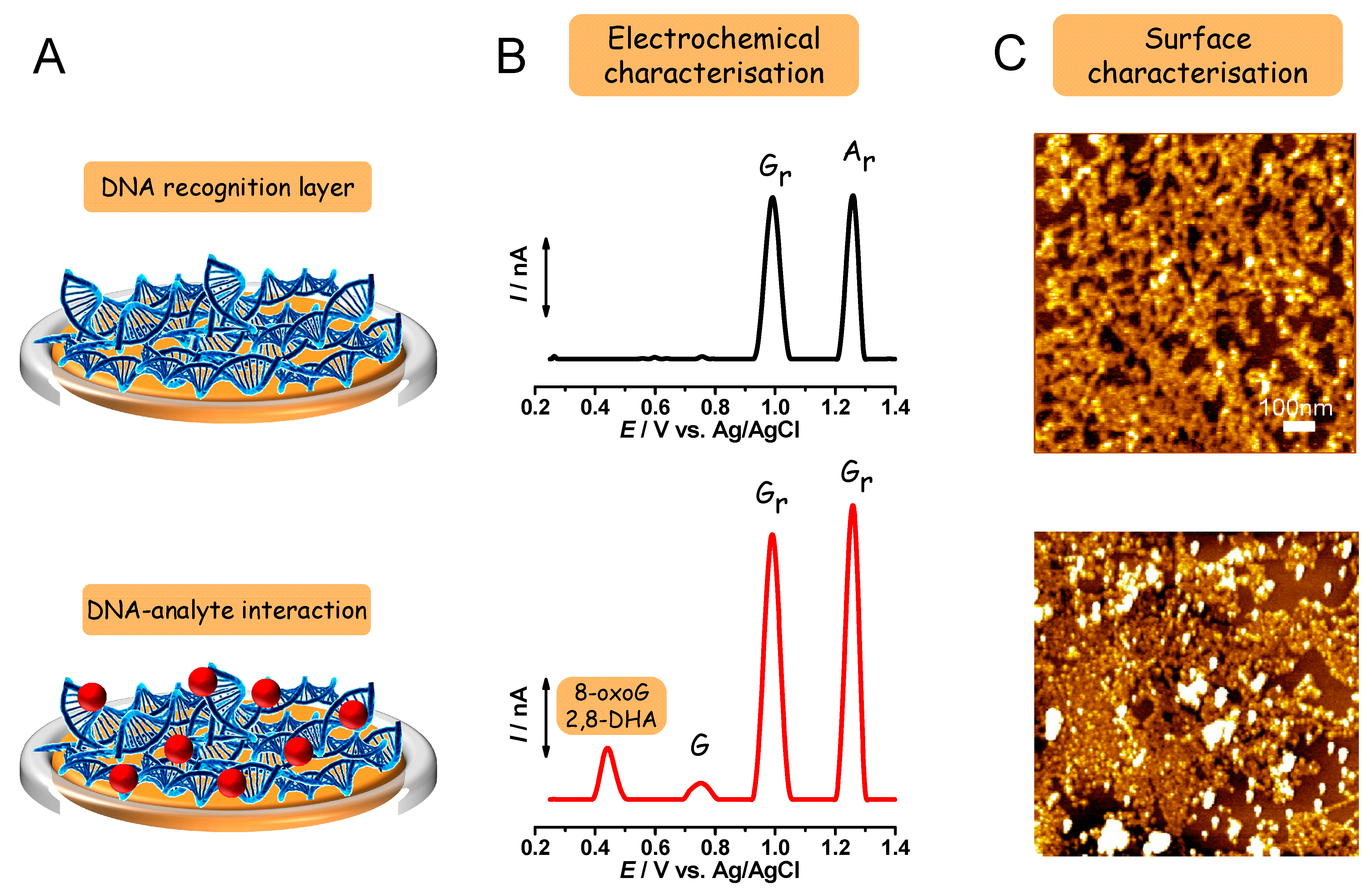
2.1. Electrochemical Characterization
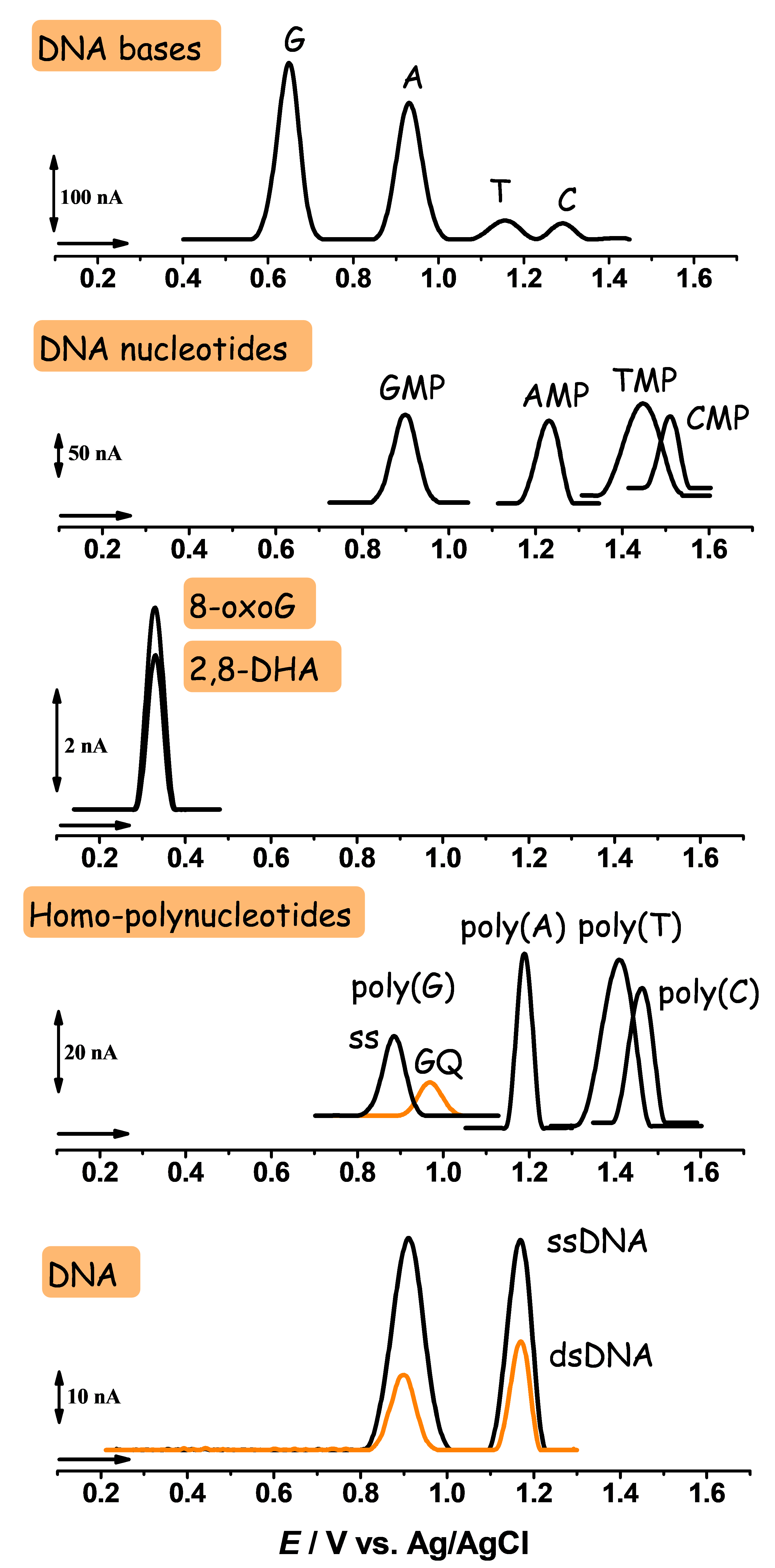
2.1.1. Electrochemical Detection of DNA Base, Nucleoside, and Nucleotide Oxidation
2.1.2. Electrochemical Detection of 8-oxodG and 2,8-DHA Biomarkers Oxidation
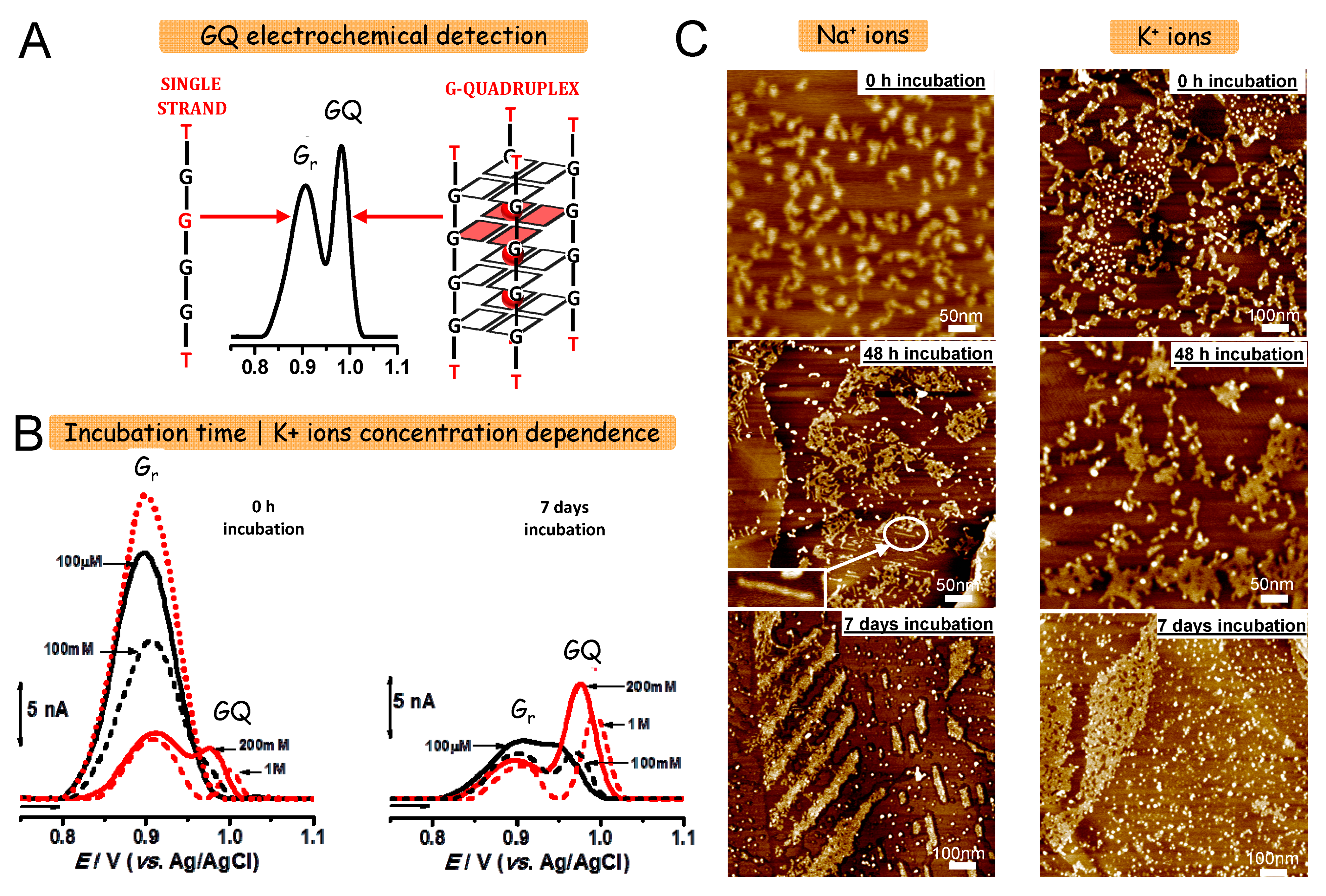
2.1.3. Electrochemical Detection of DNA Oxidation
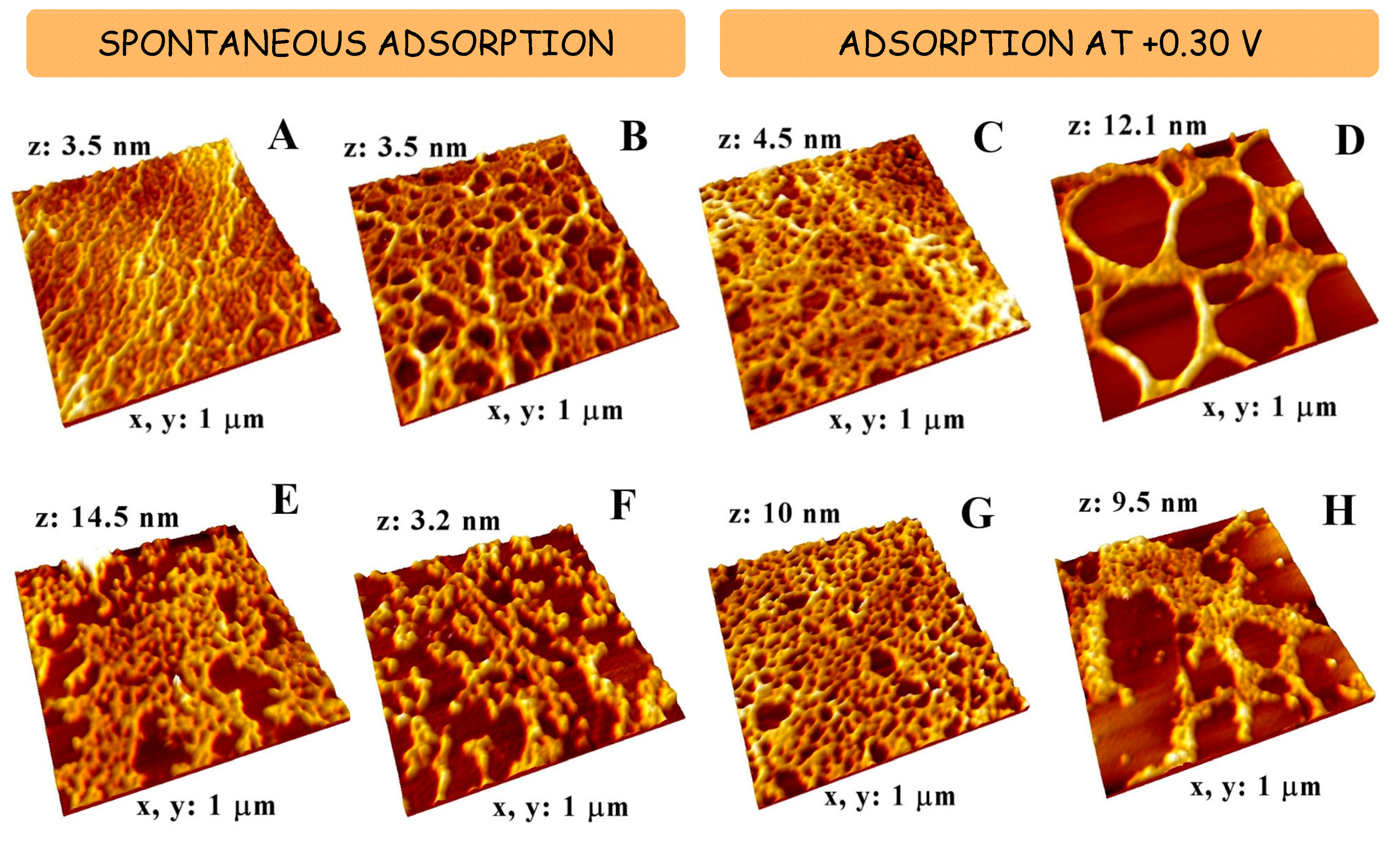
2.2. Morphological Characterization
3. DNA Electrochemical Biosensors for the Detection of Pharmaceutical Drugs
3.1. Acridine Derivatives
3.2. Alkaloids
3.3. Alkylating Agents
3.4. Alkylphosphocholines
3.5. Antibiotics
3.6. Antimetabolites
3.7. Kinase Inhibitors
3.8. Immunomodulatory Agents
3.9. Nucleoside Analogs
3.10. Metal Complexes
3.11. Phenolic Compounds
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kino, K.; Hirao-Suzuki, M.; Morikawa, M.; Sakaga, A.; Miyazawa, H. Generation, repair and replication of guanine oxidation products. Genes Environ. 2017, 39, 21. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.N.; Jain, N.; Garg, D.K. Electrochemical and enzymic oxidation of guanosine and 8-hydroxyguanosine and the effects of oxidation products in mice. Bioelectrochem. Bioenerg. 1997, 43, 105–114. [Google Scholar] [CrossRef]
- Brett, C.M.A.; Oliveira-Brett, A.M.; Serrano, S.H.P. On the adsorption and electrochemical oxidation of DNA at glassy carbon electrodes. J. Electroanal. Chem. 1994, 366, 225–231. [Google Scholar] [CrossRef]
- Goyal, R.; Dryhurst, G. Redox chemistry of guanine and 8-oxyguanine and a comparison of the peroxidase-catalyzed and electrochemical oxidation of 8-oxyguanine. J. Electroanal. Chem. Interfacial Electrochem. 1982, 135, 75–91. [Google Scholar] [CrossRef]
- Yudkina, A.V.; Shilkin, E.S.; Endutkin, A.V.; Makarova, A.V.; Zharkov, D.O. Reading and Misreading 8-oxoguanine, a Paradigmatic Ambiguous Nucleobase. Crystals 2019, 9, 269. [Google Scholar] [CrossRef] [Green Version]
- Loft, S.; Danielsen, P.; Løhr, M.; Jantzen, K.; Hemmingsen, J.G.; Roursgaard, M.; Karotki, D.G.; Møller, P. Urinary excretion of 8-oxo-7,8-dihydroguanine as biomarker of oxidative damage to DNA. Arch. Biochem. Biophys. 2012, 518, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Labuda, J.; Oliveira-Brett, A.M.; Evtugyn, G.; Fojta, M.; Mascini, M.; Ozsoz, M.; Palchetti, I.; Paleček, E.; Wang, J. Electrochemical nucleic acid-based biosensors: Concepts, terms, and methodology (IUPAC Technical Report). Pure Appl. Chem. 2010, 82, 1161–1187. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.; Serrano, S.H.P.; Piedade, A.J.P. Electrochemistry of DNA. In Applications of Kinetic Modelling; Compton, R.G., Ed.; Comprehensive Chemical Kinetics; Elsevier: Amsterdam, The Netherlands, 1999; Volume 37, pp. 91–119. ISBN 9780444501646. [Google Scholar]
- Oliveira-Brett, A.M. DNA-based biosensors. In Biosensors and Modern Biospecific Analytical Techniques; L. Gorton, Ed.; Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2005; Volume 44, pp. 179–208. ISBN 9780444507150. [Google Scholar]
- Oliveira-Brett, A.M.; Chiorcea-Paquim, A.; Diculescu, V.C.; Piedade, J. Electrochemistry of nanoscale DNA surface films on carbon. Med Eng. Phys. 2006, 28, 963–970. [Google Scholar] [CrossRef] [Green Version]
- Chiorcea-Paquim, A.-M.; Oliveira, S.C.; Diculescu, V.C.; Oliveira-Brett, A.M. Applications of DNA-Electrochemical Biosensors in Cancer Research. In Comprehensive Analytical Chemistry; Barcelo, D., Hansen, P.-D., Palchetti, I., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; Volume 77, pp. 287–336. ISBN 9780444639462. [Google Scholar]
- Oliveira-Brett, A.M. Electrochemistry for probing DNA damage. In Encyclopedia of Sensors; Grimes, C.A., Dickey, E.C., Pishko, M.V., Eds.; American Scientific Publishers: Stevenson Ranch, CA, USA, 2006; p. 301. ISBN 1-58883-056-X. [Google Scholar]
- Oliveira-Brett, A.M.; Diculescu, V.C.; Chiorcea-Paquim, A.M.; Serrano, S.H.P. Electrochemical Sensor Analysis. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2007; Volume 49, pp. 413–437. ISBN 9780444530530. [Google Scholar]
- Oliveira-Brett, A.M. Electrochemical DNA Assays. In Bioelectrochemistry: Fundamentals, Experimental Techniques and Applications; Bartlett, P.N., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2008; p. 411. ISBN 9780470753842. [Google Scholar]
- Oliveira-Brett, A.-M. Nanobioelectrochemistry. In Electrochemistry at the Nanoscale, Nanostrutures Science and Technology; Schmuki, P., Virtanen, S., Eds.; Nanostructure Science and Technology; Springer: New York, NY, USA, 2009; p. 407. ISBN 978-0-387-73581-8. [Google Scholar]
- Oliveira-Brett, A.M.; Diculescu, V.C.; Chiorcea-Paquim, A.; Serrano, S.H.P. Chapter 20 DNA-electrochemical biosensors for investigating DNA damage. In Vibrational Spectroscopy for Plant Varieties and Cultivars Characterization; Elsevier BV: Amsterdam, The Netherlands, 2007; Volume 49, pp. 413–437. [Google Scholar]
- Diculescu, V.C.; Chiorcea-Paquim, A.; Oliveira-Brett, A.M. Electrochemical DNA Sensors for Detection of DNA Damage. Sensors 2005, 5, 377–393. [Google Scholar] [CrossRef]
- Diculescu, V.C.; Chiorcea-Paquim, A.-M.; Oliveira-Brett, A.M. Applications of a DNA-electrochemical biosensor. TrAC Trends Anal. Chem. 2016, 79, 23–36. [Google Scholar] [CrossRef]
- Oliveira, S.C.B.; Diculescu, V.; Chiorcea-Paquim, A.M.; Oliveira-Brett, A. Electrochemical Biosensors for DNA–Drug Interactions. In Encyclopedia of Interfacial Chemistry; Elsevier BV: Amsterdam, The Netherlands, 2018; pp. 124–139. [Google Scholar]
- Lopes, I.C.; Oliveira-Brett, A.M. Human Cytochrome P450 (CYP1A2)-dsDNA Interactionin situ Evaluation Using a dsDNA-electrochemical Biosensor. Electroanalysis 2017, 29, 1674–1682. [Google Scholar] [CrossRef]
- Diculescu, V.C.; Chiorcea-Paquim, A.; Eritja, R.; Oliveira-Brett, A.M. Evaluation of the structure–activity relationship of thrombin with thrombin binding aptamers by voltammetry and atomic force microscopy. J. Electroanal. Chem. 2011, 656, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Chiorcea-Paquim, A.-M.; Santos, P.V.; Eritja, R.; Oliveira-Brett, A.M. Self-assembled G-quadruplex nanostructures: AFM and voltammetric characterization. Phys. Chem. Chem. Phys. 2013, 15, 9117–9124. [Google Scholar] [CrossRef]
- Oliveira, S.C.B.; Oliveira-Brett, A.M. In situ evaluation of chromium–DNA damage using a DNA-electrochemical biosensor. Anal. Bioanal. Chem. 2010, 398, 1633–1641. [Google Scholar] [CrossRef]
- Oliveira, S.C.B.; Corduneanu, O.; Oliveira-Brett, A.M. In situ evaluation of heavy metal–DNA interactions using an electrochemical DNA biosensor. Bioelectrochemistry 2008, 72, 53–58. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.; Corduneanu, O.; Oliveira, S.C.B.; Diculescu, V.C.; Oliveira-Brett, A.M. Electrochemical and AFM evaluation of hazard compounds–DNA interaction. Electrochim. Acta 2009, 54, 1978–1985. [Google Scholar] [CrossRef]
- Lopes, I.C.; Santos, P.V.F.; Diculescu, V.C.; Peixoto, F.M.P.; Araújo, M.C.U.; Tanaka, A.A.; Oliveira-Brett, A.M. Microcystin-LR and chemically degraded microcystin-LR electrochemical oxidation. Analyst 2012, 137, 1904–1912. [Google Scholar] [CrossRef]
- Santos, P.V.F.; Lopes, I.C.; Diculescu, V.C.; Oliveira-Brett, A.M. DNA–Cyanobacterial Hepatotoxins Microcystin-LR and Nodularin Interaction: Electrochemical Evaluation. Electroanalysis 2012, 24, 547–553. [Google Scholar] [CrossRef]
- Enache, T.A.; Chiorcea-Paquim, A.-M.; Fatibello-Filho, O.; Oliveira-Brett, A.M. Hydroxyl radicals electrochemically generated in situ on a boron-doped diamond electrode. Electrochem. Commun. 2009, 11, 1342–1345. [Google Scholar] [CrossRef]
- Oliveira, S.C.B.; Oliveira-Brett, A.M. Boron doped diamond electrode pre-treatments effect on the electrochemical oxidation of dsDNA, DNA bases, nucleotides, homopolynucleotides and biomarker 8-oxoguanine. J. Electroanal. Chem. 2010, 648, 60–66. [Google Scholar] [CrossRef]
- Oliveira, S.C.B.; Oliveira-Brett, A.M. In Situ DNA Oxidative Damage by Electrochemically Generated Hydroxyl Free Radicals on a Boron-Doped Diamond Electrode. Langmuir 2012, 28, 4896–4901. [Google Scholar] [CrossRef]
- Piedade, J.A.P.; Oliveira, P.S.C.; Lopes, M.C.; Oliveira-Brett, A.M. Voltammetric determination of γ radiation-induced DNA damage. Anal. Biochem. 2006, 355, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Brett, A.M.; Piedade, J.A.P.; Silva, L.A.; Diculescu, V. Voltammetric determination of all DNA nucleotides. Anal. Biochem. 2004, 332, 321–329. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, S.C.B.; Oliveira-Brett, A.M. DNA-electrochemical biosensors: AFM surface characterisation and application to detection of in situ oxidative damage to DNA. Comb. Chem. High Throughput Screen. 2010, 13, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Chiorcea-Paquim, A.M.; Pontinha, A.D.R.; Oliveira-Brett, A.M. Time-dependent polyguanylic acid structural modifications. Electrochem. Commun. 2014, 45, 71–74. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.; Piedade, J.A.P.; Serrano, S.H.P. Electrochemical Oxidation of 8-Oxoguanine. Electroanalalysis 2000, 12, 969–973. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.; Diculescu, V.; Piedade, J. Electrochemical oxidation mechanism of guanine and adenine using a glassy carbon microelectrode. Bioelectrochemistry 2002, 55, 61–62. [Google Scholar] [CrossRef] [Green Version]
- David, S.S.; O’Shea, V.L.; Kundu, S. Base-excision repair of oxidative DNA damage. Nat. Cell Biol. 2007, 447, 941–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkateswarlu, D.; Leszczynski, J. Tautomeric equilibria in 8-oxopurines: Implications for mutagenicity. J. Comput. Mol. Des. 1998, 12, 373. [Google Scholar] [CrossRef] [PubMed]
- Drake, D.M.; Shapiro, A.M.; Wells, P.G. Measurement of the Oxidative DNA Lesion 8-Oxoguanine (8-oxoG) by ELISA or by High-Performance Liquid Chromatography (HPLC) with Electrochemical Detection. Breast Cancer 2019, 1965, 313–328. [Google Scholar] [CrossRef]
- Valavanidis, A.; Vlachogianni, T.; Fiotakis, C. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): A Critical Biomarker of Oxidative Stress and Carcinogenesis. J. Environ. Sci. Health Part C 2009, 27, 120–139. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, P.; Dryhurst, G. Electrochemical oxidation of guanosine formation of some novel guanine oligonucleosides. J. Electroanal. Chem. Interfacial Electrochem. 1987, 224, 137–162. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.; Matysik, F.-M. Sonoelectrochemical studies of guanine and guanosine. Bioelectrochem. Bioenerg. 1997, 42, 111–116. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.; Da Silva, L.A.; Brett, C.M.A. Adsorption of Guanine, Guanosine, and Adenine at Electrodes Studied by Differential Pulse Voltammetry and Electrochemical Impedance. Langmuir 2002, 18, 2326–2330. [Google Scholar] [CrossRef] [Green Version]
- Chiorcea, A.-M.; Oliveira-Brett, A.M. Scanning probe microscopic imaging of guanine on a highly oriented pyrolytic graphite electrode. Bioelectrochemistry 2002, 55, 63–65. [Google Scholar] [CrossRef] [Green Version]
- Oliveira-Brett, A.M.; Brett, C.M.A.; Silva, L. An impedance study of the adsorption of nucleic acid bases at glassy carbon electrodes. Bioelectrochemistry 2002, 56, 33–35. [Google Scholar] [CrossRef] [Green Version]
- Rebelo, I.; Piedade, J.A.P.; Oliveira-Brett, A.M. Electrochemical determination of 8-oxoguanine in the presence of uric acid. Bioelectrochemistry 2004, 63, 267–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, P.; Oyama, M.; Goyal, R.N. Electrochemical investigations of 8-hydroxydeoxyguanosine and its determination at an edge plane pyrolytic graphite electrode. RSC Adv. 2015, 6, 1722–1728. [Google Scholar] [CrossRef]
- Langmaier, J.; Samec, Z.; Samcová, E. Electrochemical Oxidation of 8-Oxo-2′-Deoxyguanosine on Glassy Carbon, Gold, Platinum and Tin(IV) Oxide Electrodes. Electroanalysis 2003, 15, 1555–1560. [Google Scholar] [CrossRef]
- Shibata, J.H. Nucleic Acids: Structures, Properties, and Functions (Bloomfield, Victor A.; Crothers, Donald M.; Tinoco, Ignacio, Jr.; contributions from Hearst, John E.; Wemmer, David E.; Kollman, Peter A.; Turner, Douglas H.). J. Chem. Educ. 2001, 78, 314. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.; Santos, P.V.; Oliveira-Brett, A.M. Atomic force microscopy and voltammetric characterisation of synthetic homo-oligodeoxynucleotides. Electrochim. Acta 2013, 110, 599–607. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.-M.; Oliveira-Brett, A.M. Guanine Quadruplex Electrochemical Aptasensors. Chemosensors 2016, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Chiorcea-Paquim, A.-M.; Oliveira-Brett, A.M. Redox behaviour of G-quadruplexes. Electrochim. Acta 2014, 126, 162–170. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.; Santos, P.; Diculescu, V.; Eritja, R.; Oliveira-Brett, A. Electrochemical Characterization of Guanine Quadruplexes. In Guanine Quartets; Royal Society of Chemistry (RSC): London, UK, 2012; pp. 100–109. [Google Scholar]
- Diculescu, V.C.; Chiorcea-Paquim, A.; Eritja, R.; Oliveira-Brett, A.M. Thrombin-Binding Aptamer Quadruplex Formation: AFM and Voltammetric Characterization. J. Nucleic Acids 2010, 2010, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontinha, A.D.R.; Chiorcea-Paquim, A.; Eritja, R.; Oliveira-Brett, A.M. Quadruplex Nanostructures of d(TGGGGT): Influence of Sodium and Potassium Ions. Anal. Chem. 2014, 86, 5851–5857. [Google Scholar] [CrossRef]
- Chiorcea, A.-M.; Oliveira-Brett, A.M. Atomic force microscopy characterization of an electrochemical DNA-biosensor. Bioelectrochemistry 2004, 63, 229–232. [Google Scholar] [CrossRef] [Green Version]
- Oliveira-Brett, A.M.; Chiorcea, A.-M. Effect of pH and applied potential on the adsorption of DNA on highly oriented pyrolytic graphite electrodes. Atomic force microscopy surface characterisation. Electrochem. Commun. 2003, 5, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Chiorcea-Paquim, A.; Piedade, J.A.P.; Wombacher, R.; Jäschke, A.; Oliveira-Brett, A.M. Atomic Force Microscopy and Anodic Voltammetry Characterization of a 49-Mer Diels-Alderase Ribozyme. Anal. Chem. 2006, 78, 8256–8264. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.; Chiorcea, A.-M. Atomic Force Microscopy of DNA Immobilized onto a Highly Oriented Pyrolytic Graphite Electrode Surface. Langmuir 2003, 19, 3830–3839. [Google Scholar] [CrossRef] [Green Version]
- Chiorcea-Paquim, A.; Diculescu, V.C.; Oretskaya, T.; Oliveira-Brett, A.M. AFM and electroanalytical studies of synthetic oligonucleotide hybridization. Biosens. Bioelectron. 2004, 20, 933–944. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.; Chiorcea-Paquim, A.-M. DNA imaged on a HOPG electrode surface by AFM with controlled potential. Bioelectrochemistry 2005, 66, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Brett, A.M.; Chiorcea-Paquim, A.M.; Diculescu, V.C.; Oretskaya, T. Synthetic oligonucleotides: AFM characterisation and electroanalytical studies. Bioelectrochemistry 2005, 67, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Chiorcea-Paquim, A.; Oretskaya, T.S.; Oliveira-Brett, A.M. Adsorption of synthetic homo- and hetero-oligodeoxynucleotides onto highly oriented pyrolytic graphite: Atomic force microscopy characterization. Biophys. Chem. 2006, 121, 131–141. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.; Oretskaya, T.S.; Oliveira-Brett, A.M. Atomic force microscopy characterization of synthetic pyrimidinic oligodeoxynucleotides adsorbed onto an HOPG electrode under applied potential. Electrochim. Acta 2006, 51, 5037–5045. [Google Scholar] [CrossRef]
- Tadini-Buoninsegni, F.; Palchetti, I. Label-Free Bioelectrochemical Methods for Evaluation of Anticancer Drug Effects at a Molecular Level. Sensors 2020, 20, 1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiorcea-Paquim, A.; Pontinha, A.D.R.; Oliveira-Brett, A.M. Quadruplex-targeting anticancer drug BRACO-19 voltammetric and AFM characterization. Electrochim. Acta 2015, 174, 155–163. [Google Scholar] [CrossRef]
- Pontinha, A.D.R.; Sparapani, S.; Neidle, S.; Oliveira-Brett, A.M. Triazole–acridine conjugates: Redox mechanisms and in situ electrochemical evaluation of interaction with double-stranded DNA. Bioelectrochemistry 2013, 89, 50–56. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.; Pontinha, A.D.R.; Eritja, R.; Lucarelli, G.; Sparapani, S.; Neidle, S.; Oliveira-Brett, A.M. Atomic Force Microscopy and Voltammetric Investigation of Quadruplex Formation between a Triazole-Acridine Conjugate and Guanine-Containing Repeat DNA Sequences. Anal. Chem. 2015, 87, 6141–6149. [Google Scholar] [CrossRef] [Green Version]
- Javar, H.A.; Garkani-Nejad, Z.; Noudeh, G.D.; Mahmoudi-Moghaddam, H. Development of a new electrochemical DNA biosensor based on Eu3+-doped NiO for determination of amsacrine as an anti-cancer drug: Electrochemical, spectroscopic and docking studies. Anal. Chim. Acta 2020, 1133, 48–57. [Google Scholar] [CrossRef]
- Untiveros, K.L.; Da Silva, E.G.; De Abreu, F.C.; Da Silva-Júnior, E.F.; De Araújo-Junior, J.X.; De Aquino, T.M.; Armas, S.M.; De Moura, R.O.; Mendonça-Junior, F.J.; Serafim, V.L.; et al. An electrochemical biosensor based on Hairpin-DNA modified gold electrode for detection of DNA damage by a hybrid cancer drug intercalation. Biosens. Bioelectron. 2019, 133, 160–168. [Google Scholar] [CrossRef]
- Mahmoudi-Moghaddam, H.; Tajik, S.; Beitollahi, H. A new electrochemical DNA biosensor based on modified carbon paste electrode using graphene quantum dots and ionic liquid for determination of topotecan. Microchem. J. 2019, 150, 104085. [Google Scholar] [CrossRef]
- Javar, H.A.; Mahmoudi-Moghaddam, H. A Label-Free DNA Biosensor for Determination of Topotecan as an Anticancer Drug: Electrochemical, Spectroscopic and Docking Studies. J. Electrochem. Soc. 2020, 167, 127502. [Google Scholar] [CrossRef]
- Bolat, G. Investigation of poly(CTAB-MWCNTs) composite based electrochemical DNA biosensor and interaction study with anticancer drug Irinotecan. Microchem. J. 2020, 159, 105426. [Google Scholar] [CrossRef]
- Vajedi, F.S.; Dehghani, H. A high-sensitive electrochemical DNA biosensor based on a novel ZnAl/layered double hydroxide modified cobalt ferrite-graphene oxide nanocomposite electrophoretically deposited onto FTO substrate for electroanalytical studies of etoposide. Talanta 2020, 208, 120444. [Google Scholar] [CrossRef]
- Ghalkhani, M.; Fernandes, I.P.G.; Oliveira, S.C.B.; Shahrokhianab, S.; Oliveira-Brett, A.M. Electrochemical Redox Behaviour of Temozolomide Using a Glassy Carbon Electrode. Electroanalysis 2010, 22, 2633–2640. [Google Scholar] [CrossRef]
- Lopes, I.C.; De Oliveira, S.C.B.; Oliveira-Brett, A.M. Temozolomide chemical degradation to 5-aminoimidazole-4-carboxamide – Electrochemical study. J. Electroanal. Chem. 2013, 704, 183–189. [Google Scholar] [CrossRef]
- Lopes, I.C.; Oliveira, S.C.B.; Oliveira-Brett, A.M. In situ electrochemical evaluation of anticancer drug temozolomide and its metabolites–DNA interaction. Anal. Bioanal. Chem. 2012, 405, 3783–3790. [Google Scholar] [CrossRef]
- Jahandari, S.; Taher, M.A.; Karimi-Maleh, H.; Khodadadi, A.; Faghih-Mirzaei, E. A powerful DNA-based voltammetric biosensor modified with Au nanoparticles, for the determination of Temodal; an electrochemical and docking investigation. J. Electroanal. Chem. 2019, 840, 313–318. [Google Scholar] [CrossRef]
- De Carvalho, P.A.V.; Lopes, I.C.; Silva, E.H.C.; Bruzaca, E.E.S.; Alves, H.J.; Lima, M.I.S.; Tanaka, A.A.; Chaves, H.J. Electrochemical behaviour of anticancer drug lomustine and in situ evaluation of its interaction with DNA. J. Pharm. Biomed. Anal. 2019, 176, 112786. [Google Scholar] [CrossRef] [PubMed]
- Machini, W.B.S.; Oliveira-Brett, A.M. Antileishmanial Drug Miltefosine-dsDNA Interaction in situ Evaluation with a DNA-Electrochemical Biosensor. Electroanalysis 2018, 30, 48–56. [Google Scholar] [CrossRef]
- Hortobágyi, G.N. Anthrazykline in der Krebstherapie. Drugs 1997, 54, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Piedade, J.; Fernandes, I.; Oliveira-Brett, A.M. Electrochemical sensing of DNA–adriamycin interactions. Bioelectrochemistry 2002, 56, 81–83. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.; Piedade, J.A.P.; Chiorcea, A.-M. Anodic voltammetry and AFM imaging of picomoles of adriamycin adsorbed onto carbon surfaces. J. Electroanal. Chem. 2002, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Oliveira-Brett, A.M.; Vivan, M.; Fernandes, I.R.; Piedade, J.A.P. Electrochemical detection of in situ adriamycin oxidative damage to DNA. Talanta 2002, 56, 959–970. [Google Scholar] [CrossRef] [Green Version]
- Kulikova, T.; Porfir’Eva, A.V.; Evtugyn, G.; Hianik, T. Electrochemical DNA Sensors with Layered Polyaniline—DNA Coating for Detection of Specific DNA Interactions. Sensors 2019, 19, 469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kara, H.E. Şatana Redox mechanism of anticancer drug idarubicin and in-situ evaluation of interaction with DNA using an electrochemical biosensor. Bioelectrochemistry 2014, 99, 17–23. [Google Scholar] [CrossRef]
- Congur, G.; Eksin, E.; Erdem, A. Levan modified DNA biosensor for voltammetric detection of daunorubicin-DNA interaction. Sensors Actuators B Chem. 2021, 326, 128818. [Google Scholar] [CrossRef]
- Khodadadi, A.; Faghih-Mirzaei, E.; Karimi-Maleh, H.; Abbaspourrad, A.; Agarwal, S.; Gupta, V.K. A new epirubicin biosensor based on amplifying DNA interactions with polypyrrole and nitrogen-doped reduced graphene: Experimental and docking theoretical investigations. Sensors Actuators B Chem. 2019, 284, 568–574. [Google Scholar] [CrossRef]
- He, X.; Han, H.; Shi, W.; Dong, J.; Lu, X.; Yang, W.; Lu, X. A label-free electrochemical DNA biosensor for kanamycin detection based on diblock DNA with poly-cytosine as a high affinity anchor on graphene oxide. Anal. Methods 2020, 12, 3462–3469. [Google Scholar] [CrossRef] [PubMed]
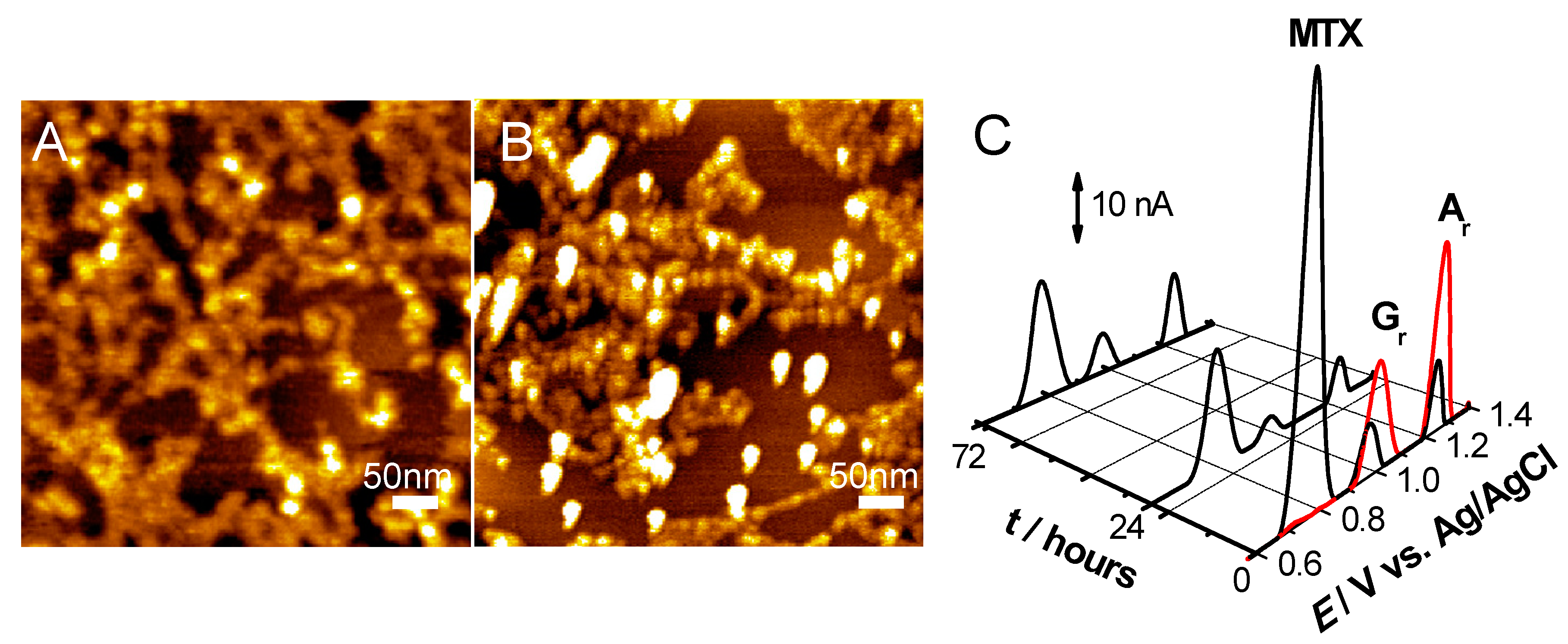
- Pontinha, A.D.R.; Jorge, S.M.A.; Paquim, A.-M.C.; Diculescu, V.C.; Oliveira-Brett, A.M. In situ evaluation of anticancer drug methotrexate–DNA interaction using a DNA-electrochemical biosensor and AFM characterization. Phys. Chem. Chem. Phys. 2011, 13, 5227–5234. [Google Scholar] [CrossRef]
- Queiroz, N.L.; Nascimento, M.L.; Nascimento, J.A.M.; Nascimento, V.B.; Oliveira, S.C.B. Electrochemistry Study of Antineoplastic Raltitrexed Oxidation Mechanism and its Interaction with DNA. Electroanalysis 2018, 30, 1184–1191. [Google Scholar] [CrossRef]
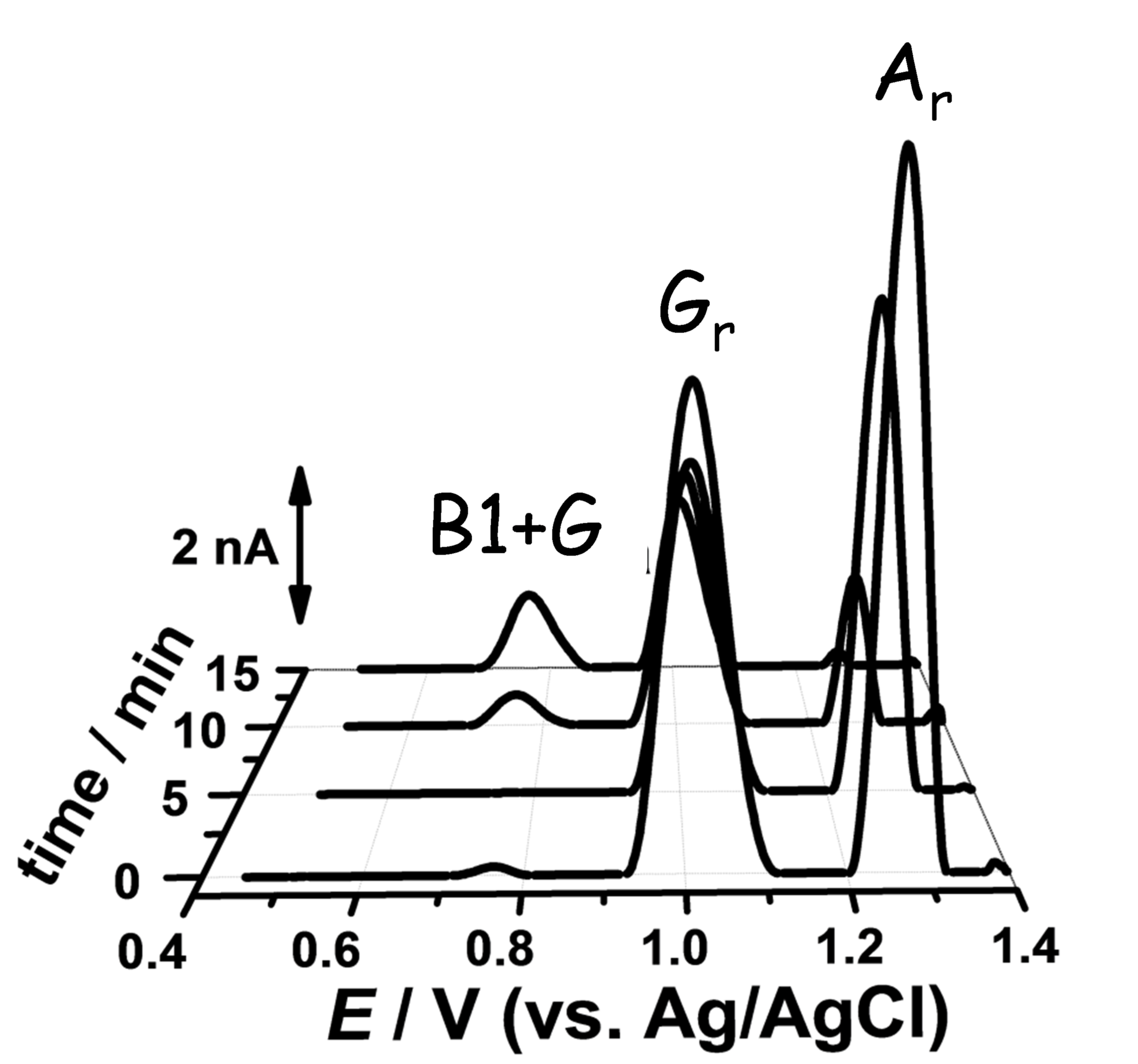
- Diculescu, V.C.; Vivan, M.; Oliveira-Brett, A.M. Voltammetric Behavior of Antileukemia Drug Glivec. Part I–Electrochemical Study of Glivec. Electroanalysis 2006, 18, 1800–1807. [Google Scholar] [CrossRef] [Green Version]
- Diculescu, V.C.; Vivan, M.; Oliveira-Brett, A.M. Voltammetric Behavior of Antileukemia Drug Glivec. Part II—Redox Processes of Glivec Electrochemical Metabolite. Electroanalysis 2006, 18, 1808–1814. [Google Scholar] [CrossRef] [Green Version]
- Diculescu, V.C.; Vivan, M.; Oliveira-Brett, A.M. Voltammetric Behavior of Antileukemia Drug Glivec. Part III: In Situ DNA Oxidative Damage by the Glivec Electrochemical Metabolite. Electroanalysis 2006, 18, 1963–1970. [Google Scholar] [CrossRef] [Green Version]
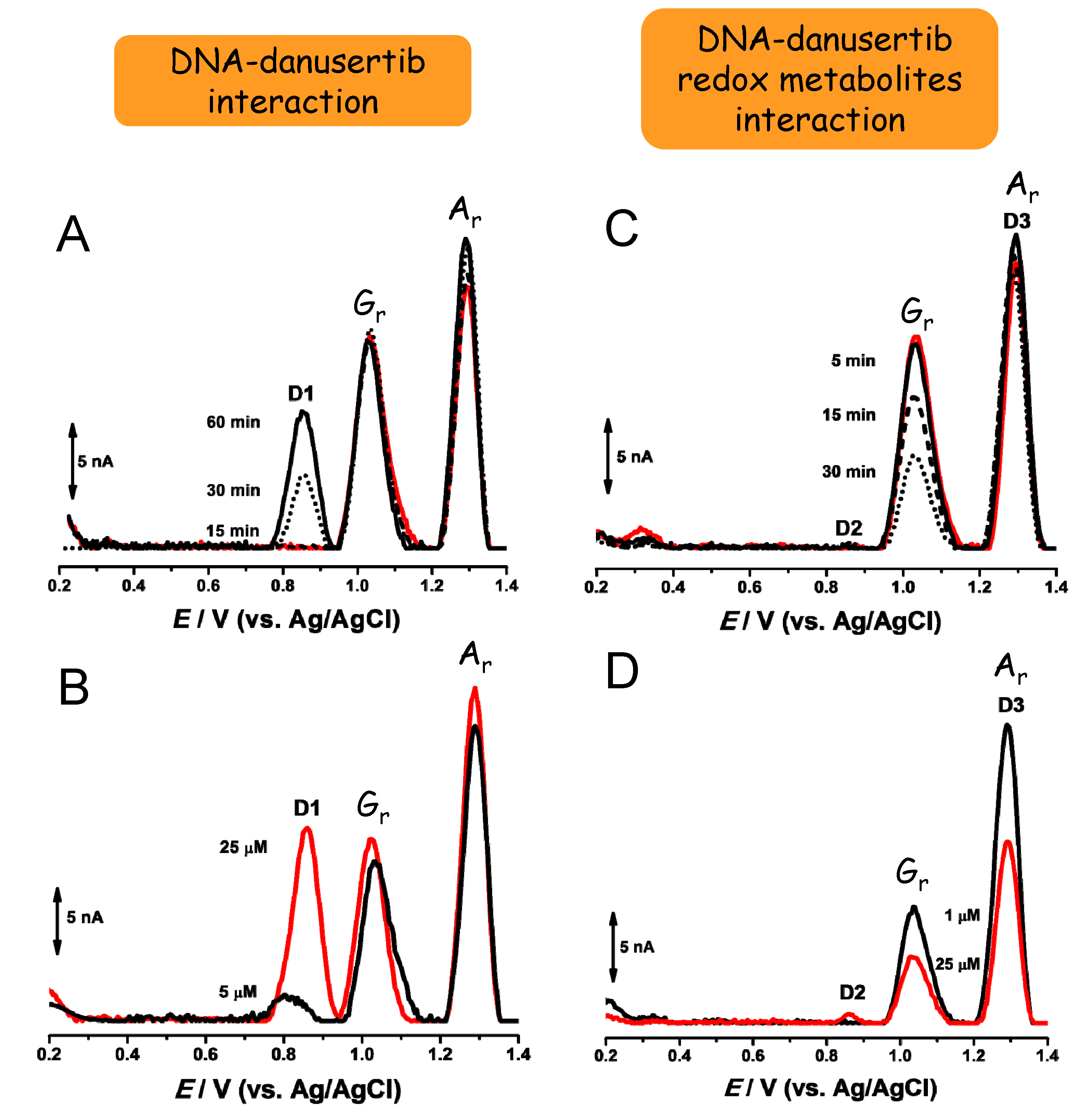
- Diculescu, V.C.; Oliveira-Brett, A.M. In situ electrochemical evaluation of dsDNA interaction with the anticancer drug danusertib nitrenium radical product using the DNA-electrochemical biosensor. Bioelectrochemistry 2016, 107, 50–57. [Google Scholar] [CrossRef]
- Topal, B.D.; Bozal-Palabiyik, B.; Ozkan, S.A.; Uslu, B. Investigation of anticancer drug lapatinib and its interaction with dsDNA by electrochemical and spectroscopic techniques. Sensors Actuators B Chem. 2014, 194, 185–194. [Google Scholar] [CrossRef]
- Oliveira, S.C.B.; Chiorcea-Paquim, A.; Ribeiro, S.; Melo, A.; Vivan, M.; Oliveira-Brett, A.M. In situ electrochemical and AFM study of thalidomide–DNA interaction. Bioelectrochemistry 2009, 76, 201–207. [Google Scholar] [CrossRef]
- Machini, W.B.S.; Marques, N.V.; Oliveira-Brett, A.M. In Situ Evaluation of Anticancer Monoclonal Antibody Nivolumab-DNA Interaction Using a DNA-Electrochemical Biosensor. ChemElectroChem 2019, 6, 4608–4616. [Google Scholar] [CrossRef]
- Oliveira, S.C.B.; Santarino, I.; Oliveira-Brett, A.M. Direct Electrochemistry of Native and Denatured Anticancer Antibody Rituximab at a Glassy Carbon Electrode. Electroanalysis 2013, 25, 1029–1034. [Google Scholar] [CrossRef]
- Santarino, I.; Oliveira, S.C.B.; Oliveira-Brett, A.M. In Situ Evaluation of the Anticancer Antibody Rituximab-dsDNA Interaction Using a DNA-Electrochemical Biosensor. Electroanalysis 2014, 26, 1304–1311. [Google Scholar] [CrossRef]
- Issaad, F.Z.; Tomé, L.I.; Marques, N.V.; Mouats, C.; Diculescu, V.C.; Oliveira-Brett, A.M. Bevacizumab anticancer monoclonal antibody: Native and denatured redox behaviour. Electrochim. Acta 2016, 206, 246–253. [Google Scholar] [CrossRef]
- Tomé, L.I.; Marques, N.V.; Diculescu, V.C.; Oliveira-Brett, A.M. In situ dsDNA-bevacizumab anticancer monoclonal antibody interaction electrochemical evaluation. Anal. Chim. Acta 2015, 898, 28–33. [Google Scholar] [CrossRef]
- Satana, H.E.; Pontinha, A.D.R.; Diculescu, V.C.; Oliveira-Brett, A.M. Nucleoside analogue electrochemical behaviour and in situ evaluation of DNA–clofarabine interaction. Bioelectrochemistry 2012, 87, 3–8. [Google Scholar] [CrossRef]
- Pontinha, A.D.R.; Satana, H.E.; Diculescu, V.C.; Oliveira-Brett, A.M. Anodic Oxidation of Cladribine and In Situ Evaluation of DNA-Cladribine Interaction. Electroanalysis 2011, 23, 2651–2657. [Google Scholar] [CrossRef]
- Satana, H.E.; Oliveira-Brett, A.M. In SituEvaluation of Fludarabine-DNA Interaction Using a DNA-Electrochemical Biosensor. Int. J. Electrochem. 2011, 2011, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Buoro, R.M.; Lopes, I.C.; Diculescu, V.C.; Serrano, S.H.P.; Lemos, L.; Brett, C.M.A. In situ evaluation of gemcitabine–DNA interaction using a DNA-electrochemical biosensor. Bioelectrochemistry 2014, 99, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Corduneanu, O.; Chiorcea-Paquim, A.; Fiuza, S.M.; Marques, M.P.M.; Oliveira-Brett, A.M. Polynuclear palladium complexes with biogenic polyamines: AFM and voltammetric characterization. Bioelectrochemistry 2010, 78, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corduneanu, O.; Chiorcea-Paquim, A.-M.; Diculescu, V.; Fiuza, S.M.; Marques, M.P.M.; Oliveira-Brett, A.M. DNA Interaction with Palladium Chelates of Biogenic Polyamines Using Atomic Force Microscopy and Voltammetric Characterization. Anal. Chem. 2010, 82, 1245–1252. [Google Scholar] [CrossRef] [Green Version]
- Corduneanu, O.; Chiorcea-Paquim, A.; Garnett, M.; Oliveira-Brett, A.M. Lipoic acid–palladium complex interaction with DNA, voltammetric and AFM characterization. Talanta 2009, 77, 1843–1853. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.; Enache, T.A.; Gil, E.D.S.; Oliveira-Brett, A.M. Natural phenolic antioxidants electrochemistry: Towards a new food science methodology. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1680–1726. [Google Scholar] [CrossRef] [PubMed]
- Vasconcellos, M.C.; Costa, C.D.O.; Terto, E.G.D.S.; De Moura, M.A.F.; De Vasconcelos, C.C.; De Abreu, F.C.; De Lemos, T.L.G.; Costa-Lotufo, L.V.; Montenegro, R.C.; Goulart, M.O.F. Electrochemical, spectroscopic and pharmacological approaches toward the understanding of biflorin DNA damage effects. J. Electroanal. Chem. 2016, 765, 168–178. [Google Scholar] [CrossRef]
- Babula, P.; Vanco, J.; Krejcova, L.; Hynek, D.; Sochor, J.; Adam, V.; Trnkova, L.; Hubalek, J.; Kizek, R. Voltammetric characterization of Lawsone-Copper(II) ternary complexes and their interactions with dsDNA. Int. J. Electrochem. Sci. 2012, 7, 7349–7366. [Google Scholar]
- Oliveira-Brett, A.M.; Diculescu, V.C. Electrochemical study of quercetin–DNA interactions. Bioelectrochemistry 2004, 64, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Brett, A.M.; Diculescu, V.C. Electrochemical study of quercetin–DNA interactions: Part I. Analysis in incubated solutions. Bioelectrochemistry 2004, 64, 133–141. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiorcea-Paquim, A.-M.; Oliveira-Brett, A.M. DNA Electrochemical Biosensors for In Situ Probing of Pharmaceutical Drug Oxidative DNA Damage. Sensors 2021, 21, 1125. https://doi.org/10.3390/s21041125
Chiorcea-Paquim A-M, Oliveira-Brett AM. DNA Electrochemical Biosensors for In Situ Probing of Pharmaceutical Drug Oxidative DNA Damage. Sensors. 2021; 21(4):1125. https://doi.org/10.3390/s21041125
Chicago/Turabian StyleChiorcea-Paquim, Ana-Maria, and Ana Maria Oliveira-Brett. 2021. "DNA Electrochemical Biosensors for In Situ Probing of Pharmaceutical Drug Oxidative DNA Damage" Sensors 21, no. 4: 1125. https://doi.org/10.3390/s21041125
APA StyleChiorcea-Paquim, A.-M., & Oliveira-Brett, A. M. (2021). DNA Electrochemical Biosensors for In Situ Probing of Pharmaceutical Drug Oxidative DNA Damage. Sensors, 21(4), 1125. https://doi.org/10.3390/s21041125






