Development of a Point-of-Care Assay for HIV-1 Viral Load Using Higher Refractive Index Antibody-Coated Microbeads
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
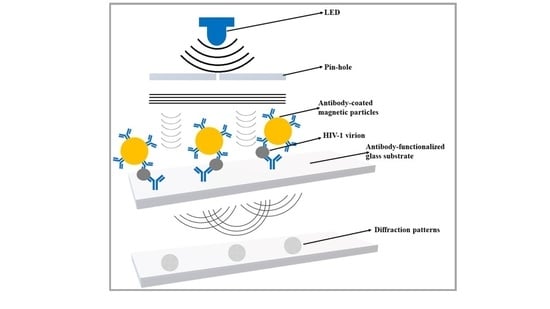
2.2.1. Design and Fabrication of Portable Lensless Imaging Setup
- (1)
- 385 nm light-emitting diode (LED) and an adjustable power supply.
- (2)
- 100-micron pinhole.
- (3)
- 18-megapixel CMOS image sensor (UI-3592LE).
- Field of View (FOV) =6.14 mm × 4.604 mm,
- Pixel size = 1.25-micron
- Overall dimensions = 60 mm × 60 mm × 138 mm (length × width × height)
2.2.2. Microchip Fabrication
2.2.3. Functionalization of Antibodies to Glass Slide
2.2.4. Viral Sample Preparation
2.2.5. Immobilization of Capture Antibody to Beads
2.2.6. HIV-1 Viral RNA Extraction, cDNA Synthesis, and Quantification Using Real-Time qPCR
2.2.7. Software
3. Results
3.1. Development of a Lensless Imaging System
3.2. Validation of the Microfluidic Chip Capture and Lensless Imaging
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- (2020, 01-18-2020). 90-90-90: Treatment for All. Available online: https://www.unaids.org/en/resources/909090 (accessed on 1 January 2021).
- Kabir, M.A.; Zilouchian, H.; Caputi, M.; Asghar, W. Advances in HIV diagnosis and monitoring. Crit. Rev. Biotechnol. 2020, 40, 623–638. [Google Scholar] [CrossRef]
- DeMarino, C.; Pleet, M.L.; Cowen, M.; Barclay, R.A.; Akpamagbo, Y.; Erickson, J.; Ndembi, N.; Charurat, M.; Jumare, J.; Bwala, S. Antiretroviral drugs alter the content of extracellular vesicles from HIV-1-infected cells. Sci. Rep. 2018, 8, 7653. [Google Scholar] [CrossRef] [PubMed]
- Broder, S. The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antivir. Res. 2010, 85, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volberding, P.A.; Deeks, S.G. Antiretroviral therapy and management of HIV infection. Lancet 2010, 376, 49–62. [Google Scholar] [CrossRef]
- Emamzadeh-Fard, S.; E Fard, S.; SeyedAlinaghi, S.; Paydary, K. Adherence to anti-retroviral therapy and its determinants in HIV/AIDS patients: A review. Infect. Disord. -Drug Targets (Former. Curr. Drug Targets-Infect. Disord.) 2012, 12, 346–356. [Google Scholar] [CrossRef] [PubMed]
- DeMarino, C.; Pleet, M.L.; Cowen, M.; Barclay, R.A.; Akpamagbo, Y.; Erickson, J.; Ndembi, N.; Charurat, M.; Jumare, J.; Bwala, S. Antiretroviral drugs alter the content of extracellular vesicles from HIV-1-infected cells. Sci. Rep. 2018, 8, 1–20. [Google Scholar]
- Pant Pai, N.; Peeling, R.W.; Smith, B.D.; Dowdy, D. Point-of-Care Tests for HIV, Related Coinfections, and Blood-Borne Infections; Hindawi: London, UK, 2014. [Google Scholar]
- Mackay, I.M.; Arden, K.E.; Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Fohlerová, Z.; Pekárek, J.; Basova, E.; Neužil, P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020, 153, 112041. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.; Lee, S.S.; Lee, C.-S.; Yamauchi, Y. Microfluidics: An untapped resource in viral diagnostics and viral cell biology. Curr. Clin. Microbiol. Rep. 2018, 5, 245–251. [Google Scholar] [CrossRef] [Green Version]
- Shafiee, H.; Asghar, W.; Inci, F.; Yuksekkaya, M.; Jahangir, M.; Zhang, M.H.; Durmus, N.G.; Gurkan, U.A.; Kuritzkes, D.R.; Demirci, U. Paper and flexible substrates as materials for biosensing platforms to detect multiple biotargets. Sci. Rep. 2015, 5, 8719. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Moon, S.; Kuritzkes, D.R.; Demirci, U. Quantum dot-based HIV capture and imaging in a microfluidic channel. Biosens. Bioelectron. 2009, 25, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.-L.; Wang, Z.-G.; Xie, H.-Y.; Liu, A.-A.; Lamb, D.C.; Pang, D.-W. Single-virus tracking: From imaging methodologies to virological applications. Chem. Rev. 2020, 120, 1936–1979. [Google Scholar] [CrossRef] [Green Version]
- Daaboul, G.G.; Freedman, D.S.; Scherr, S.M.; Carter, E.; Rosca, A.; Bernstein, D.; Mire, C.E.; Agans, K.N.; Hoenen, T.; Geisbert, T.W. Enhanced light microscopy visualization of virus particles from Zika virus to filamentous ebolaviruses. PLoS ONE 2017, 12, e0179728. [Google Scholar] [CrossRef] [PubMed]
- Daaboul, G.; Yurt, A.; Zhang, X.; Hwang, G.; Goldberg, B.; Unlu, M. High-throughput detection and sizing of individual low-index nanoparticles and viruses for pathogen identification. Nano Lett. 2010, 10, 4727–4731. [Google Scholar] [CrossRef]
- Bohren, C.F.; Huffman, D.R. Absorption and Scattering of Light by Small Particles; WileyVCH: Weinheim, Germany, 1998. [Google Scholar]
- Richert-Pöggeler, K.R.; Franzke, K.; Hipp, K.; Kleespies, R.G. Electron microscopy methods for virus diagnosis and high resolution analysis of viruses. Front. Microbiol. 2019, 9, 3255. [Google Scholar] [CrossRef] [PubMed]
- Debrick, K.S. Quantitative assay for plant viruses using serologically specific electron microscopy. Virology 1973, 56, 652–653. [Google Scholar] [CrossRef]
- Roingeard, P.; Raynal, P.I.; Eymieux, S.; Blanchard, E. Virus detection by transmission electron microscopy: Still useful for diagnosis and a plus for biosafety. Rev. Med Virol. 2019, 29, e2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biel, S.S.; Gelderblom, H.R. Diagnostic electron microscopy is still a timely and rewarding method. J. Clin. Virol. 1999, 13, 105–119. [Google Scholar] [CrossRef]
- Goldsmith, C.S.; Miller, S.E. Modern uses of electron microscopy for detection of viruses. Clin. Microbiol. Rev. 2009, 22, 552–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roingeard, P. Viral detection by electron microscopy: Past, present and future. Biol. Cell 2008, 100, 491–501. [Google Scholar] [CrossRef]
- Biel, S.S.; Madeley, D. Diagnostic virology: The need for electron microscopy: A discussion paper. J. Clin. Virol. 2001, 22, 1–9. [Google Scholar] [CrossRef]
- Ganesh, P.; Heller, T.; Chione, B.; Gumulira, J.; Gugsa, S.; Khan, S.; McGovern, S.; Nhlema, A.; Nkhoma, L.; Sacks, J.A. Near Point-of-Care HIV Viral Load: Targeted Testing at Large Facilities. J. Acquir. Immune Defic. Syndr. (1999) 2021, 86, 258. [Google Scholar] [CrossRef]
- Ilyas, S.; Simonson, A.E.; Asghar, W. Emerging Point-of-Care Technologies for Sickle Cell Disease Diagnostics. Clin. Chim. Acta 2019. [Google Scholar] [CrossRef]
- Rappa, K.; Samargia, J.; Sher, M.; Pino, J.S.; Rodriguez, H.F.; Asghar, W. Quantitative analysis of sperm rheotaxis using a microfluidic device. Microfluid. Nanofluidics 2018, 22, 100. [Google Scholar] [CrossRef]
- Asghar, W.; Sher, M.; Khan, N.S.; Vyas, J.M.; Demirci, U. Microfluidic chip for detection of fungal infections. ACS Omega 2019, 4, 7474–7481. [Google Scholar] [CrossRef] [PubMed]
- Sher, M.; Zhuang, R.; Demirci, U.; Asghar, W. based analytical devices for clinical diagnosis: Recent advances in the fabrication techniques and sensing mechanisms. Expert Rev. Mol. Diagn. 2017, 17, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.A.; Zilouchian, H.; Sher, M.; Asghar, W. Development of a Flow-Free Automated Colorimetric Detection Assay Integrated with Smartphone for Zika NS1. Diagnostics 2020, 10, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coarsey, C.; Coleman, B.; Kabir, M.A.; Sher, M.; Asghar, W. Development of a flow-free magnetic actuation platform for an automated microfluidic ELISA. RSC Adv. 2019, 9, 8159–8168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sher, M.; Asghar, W. Development of a multiplex fully automated assay for rapid quantification of CD4+ T cells from whole blood. Biosens. Bioelectron. 2019, 142, 111490. [Google Scholar] [CrossRef] [PubMed]
- Fennell, R.D.; Sher, M.; Asghar, W. Design, development, and performance comparison of wide field lensless and lens-based optical systems for point-of-care biological applications. Opt. Lasers Eng. 2021, 137, 106326. [Google Scholar] [CrossRef]
- Sharma, S.; Kabir, M.A.; Asghar, W. Lab-on-a-Chip Zika Detection with Reverse Transcription Loop-Mediated Isothermal Amplification–Based Assay for Point-of-Care Settings. Arch. Pathol. Lab. Med. 2020, 144, 1335–1343. [Google Scholar] [CrossRef]
- Ilyas, S.; Sher, M.; Du, E.; Asghar, W. Smartphone-based sickle cell disease detection and monitoring for point-of-care settings. Biosens. Bioelectron. 2020, 165, 112417. [Google Scholar] [CrossRef]
- Niskanen, I.; Räty, J.; Peiponen, K. Determination of the refractive index of microparticles by utilizing light dispersion properties of the particle and an immersion liquid. Talanta 2013, 115, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Shimobaba, T.; Weng, J.; Sakurai, T.; Okada, N.; Nishitsuji, T.; Takada, N.; Shiraki, A.; Masuda, N.; Ito, T. Computational wave optics library for C++: CWO++ library. Comput. Phys. Commun. 2012, 183, 1124–1138. [Google Scholar] [CrossRef] [Green Version]
- Pertuz, S.; Puig, D.; Garcia, M.A. Analysis of focus measure operators for shape-from-focus. Pattern Recognit. 2013, 46, 1415–1432. [Google Scholar] [CrossRef]
- Goodman, J.W. Introduction to Fourier Optics; Roberts and Company Publishers: Greenwood Village, CO, USA, 2005. [Google Scholar]
- McLeod, E.; Luo, W.; Mudanyali, O.; Greenbaum, A.; Ozcan, A. Toward giga-pixel nanoscopy on a chip: A computational wide-field look at the nano-scale without the use of lenses. Lab Chip 2013, 13, 2028–2035. [Google Scholar] [CrossRef] [Green Version]
- Mudanyali, O.; Tseng, D.; Oh, C.; Isikman, S.O.; Sencan, I.; Bishara, W.; Oztoprak, C.; Seo, S.; Khademhosseini, B.; Ozcan, A. Compact, light-weight and cost-effective microscope based on lensless incoherent holography for telemedicine applications. Lab Chip 2010, 10, 1417–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isikman, S.O.; Bishara, W.; Mavandadi, S.; Frank, W.Y.; Feng, S.; Lau, R.; Ozcan, A. Lens-free optical tomographic microscope with a large imaging volume on a chip. Proc. Natl. Acad. Sci. USA 2011, 108, 7296–7301. [Google Scholar] [CrossRef] [Green Version]
- Göröcs, Z.; Ozcan, A. On-chip biomedical imaging. IEEE Rev. Biomed. Eng. 2012, 6, 29–46. [Google Scholar] [CrossRef] [Green Version]
- Briggs, J.A.; Wilk, T.; Welker, R.; Kräusslich, H.G.; Fuller, S.D. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003, 22, 1707–1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briggs, J.A.; Simon, M.N.; Gross, I.; Kräusslich, H.-G.; Fuller, S.D.; Vogt, V.M.; Johnson, M.C. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004, 11, 672–675. [Google Scholar] [CrossRef]
- Endreß, T.; Lampe, M.; Briggs, J.A.; Kräusslich, H.-G.; Bräuchle, C.; Müller, B.; Lamb, D.C. HIV-1–cellular interactions analyzed by single virus tracing. Eur. Biophys. J. 2008, 37, 1291–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafiee, H.; Wang, S.; Inci, F.; Toy, M.; Henrich, T.J.; Kuritzkes, D.R.; Demirci, U. Emerging technologies for point-of-care management of HIV infection. Annu. Rev. Med. 2015, 66, 387–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugarinov, V.; Zvi, A.; Levy, R.; Hayek, Y.; Matsushita, S.; Anglister, J. NMR structure of an anti-gp120 antibody complex with a V3 peptide reveals a surface important for co-receptor binding. Structure 2000, 8, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Kowalski, M.; Potz, J.; Basiripour, L.; Dorfman, T.; Goh, W.C.; Terwilliger, E.; Dayton, A.; Rosen, C.; Haseltine, W.; Sodroski, J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 1987, 237, 1351–1355. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, J.; Waithe, D.; Carravilla, P.; Huarte, N.; Galiani, S.; Enderlein, J.; Eggeling, C. Envelope glycoprotein mobility on HIV-1 particles depends on the virus maturation state. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Chojnacki, J.; Eggeling, C. Super-resolution fluorescence microscopy studies of human immunodeficiency virus. Retrovirology 2018, 15, 41. [Google Scholar] [CrossRef]
- Kanakasabapathy, M.K.; Pandya, H.J.; Draz, M.S.; Chug, M.K.; Sadasivam, M.; Kumar, S.; Etemad, B.; Yogesh, V.; Safavieh, M.; Asghar, W. Rapid, label-free CD4 testing using a smartphone compatible device. Lab Chip 2017, 17, 2910–2919. [Google Scholar] [CrossRef]
- Ray, A.; Khalid, M.A.; Demčenko, A.; Daloglu, M.; Tseng, D.; Reboud, J.; Cooper, J.M.; Ozcan, A. Holographic detection of nanoparticles using acoustically actuated nanolenses. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- McArthur, C.C. Microfluidic Technologies for Capturing and Concentrating Human Immunodeficiency Virus-1 (HIV-1) Particles. Master’s Thesis, University of Kwazulu-Natal, Durban, South Africa, 2016. [Google Scholar]
- Rouet, F.; Ekouevi, D.K.; Chaix, M.-L.; Burgard, M.; Inwoley, A.; Tony, T.D.A.; Danel, C.; Anglaret, X.; Leroy, V.; Msellati, P. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 infection in a West African resource-limited setting. J. Clin. Microbiol. 2005, 43, 2709–2717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pope, M.; Haase, A.T. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nat. Med. 2003, 9, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.S.; Shaw, G.M.; McMichael, A.J.; Haynes, B.F. Acute HIV-1 infection. N. Engl. J. Med. 2011, 364, 1943–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daar, E.S.; Moudgil, T.; Meyer, R.D.; Ho, D.D. Transient high levels of viremia in patients with primary human immunodeficiency virus type 1 infection. N. Engl. J. Med. 1991, 324, 961–964. [Google Scholar] [CrossRef] [PubMed]
- Quinn, T.C. Acute primary HIV infection. JAMA 1997, 278, 58–62. [Google Scholar] [CrossRef]
- Shafiee, H.; Jahangir, M.; Inci, F.; Wang, S.; Willenbrecht, R.B.; Giguel, F.F.; Tsibris, A.M.; Kuritzkes, D.R.; Demirci, U. Acute on-chip hiv detection through label-free electrical sensing of viral nano-lysate. Small 2013, 9, 2553–2563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agbana, T.; Nijman, P.; Hoeber, M.; van Grootheest, D.; van Diepen, A.; van Lieshout, L.; Diehl, J.-C.; Verhaegen, M.; Vdovine, G. Detection of Schistosoma haematobium using lensless imaging and flow cytometry, a proof of principle study. In Proceedings of the Optical Diagnostics and Sensing XX: Toward Point-of-Care Diagnostics, San Francisco, CA, USA, 3 February 2020; p. 112470F. [Google Scholar]
- Chen, Y.-C.; Kuo, C.-S.; Wang, Z.-Z.; Lei, H.-Y.; Yang, C.-H.; Huang, C.-H. Lens-free holographic microscopy for complete blood analysis on a chip. In Proceedings of the Optical Diagnostics and Sensing XX: Toward Point-of-Care Diagnostics, San Francisco, CA, USA, 3 February 2020; p. 112470I. [Google Scholar]




| Microfluidic Chip Coupled with the Lensless Imaging Method (Virions/mL) | RT-qPCR Technique (Virions/mL) | Percentage Difference between Readings of Microfluidic Chip Coupled with Lensless Imaging Method and RT-qPCR Technique |
|---|---|---|
| 1,217,000 | 921,000 | 24.32 |
| 598,000 | 461,000 | 22.91 |
| 409,000 | 307,000 | 24.94 |
| 222,000 | 186,000 | 16.22 |
| 212,000 | 182,000 | 14.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sher, M.; Coleman, B.; Caputi, M.; Asghar, W. Development of a Point-of-Care Assay for HIV-1 Viral Load Using Higher Refractive Index Antibody-Coated Microbeads. Sensors 2021, 21, 1819. https://doi.org/10.3390/s21051819
Sher M, Coleman B, Caputi M, Asghar W. Development of a Point-of-Care Assay for HIV-1 Viral Load Using Higher Refractive Index Antibody-Coated Microbeads. Sensors. 2021; 21(5):1819. https://doi.org/10.3390/s21051819
Chicago/Turabian StyleSher, Mazhar, Benjamin Coleman, Massimo Caputi, and Waseem Asghar. 2021. "Development of a Point-of-Care Assay for HIV-1 Viral Load Using Higher Refractive Index Antibody-Coated Microbeads" Sensors 21, no. 5: 1819. https://doi.org/10.3390/s21051819