Advances in Plant Disease Detection and Monitoring: From Traditional Assays to In-Field Diagnostics
Abstract
1. Introduction
2. Global Regulatory Framework and Current Methodologies for Fighting against Epidemics
3. Innovative Technologies for Plant Pathology
3.1. Sensors Platforms for On-Field Monitoring
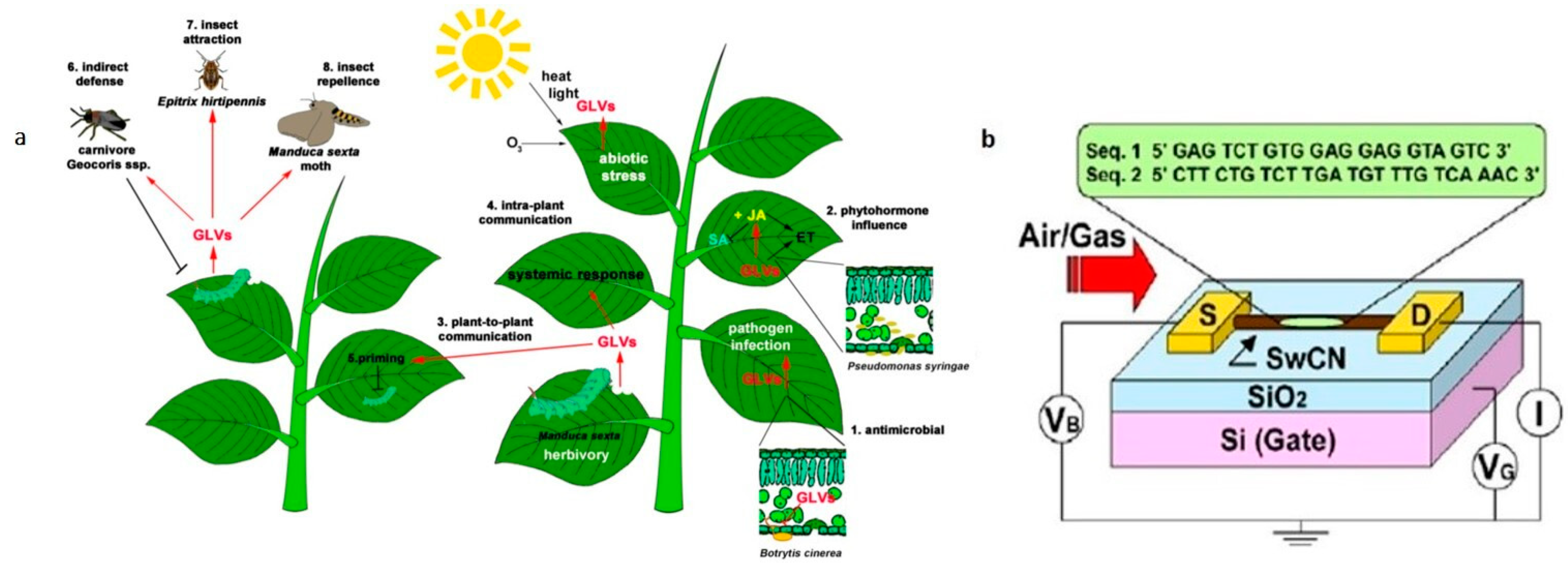
3.2. Volatile Organic Compounds Analysis for Pathogen Detection
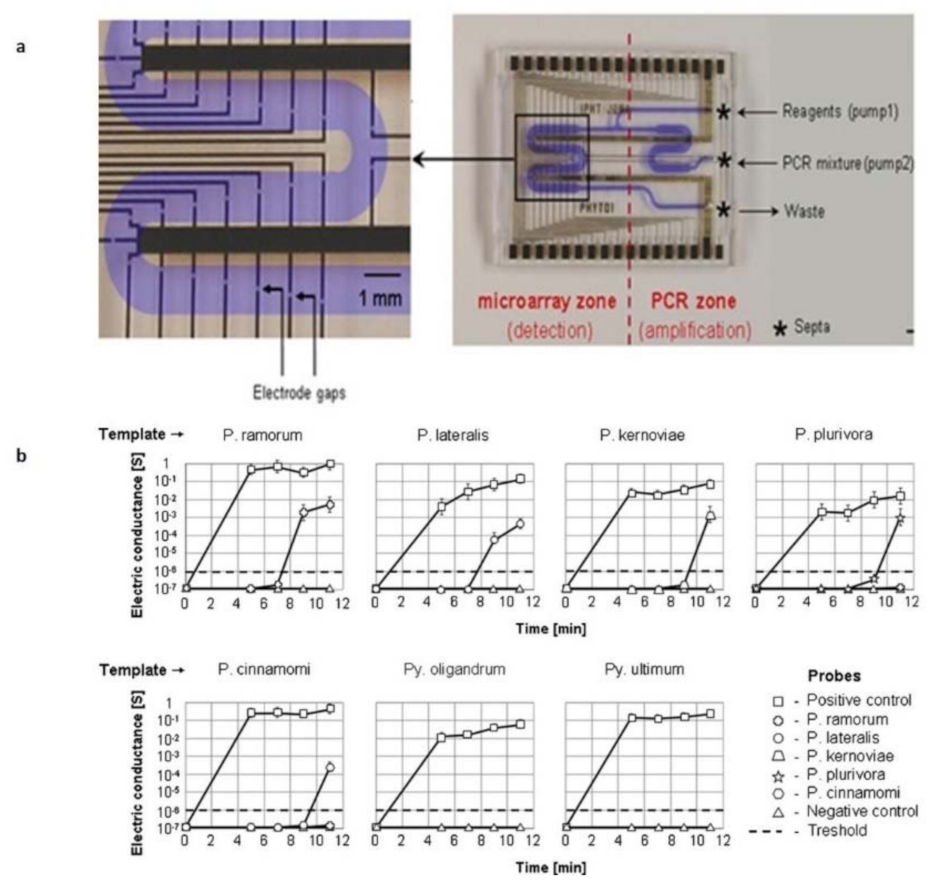
3.3. Microfluidic-Based Devices for Plant Pathogen Applications
3.4. Wearable Sensors and Their Support in Real-Time Monitoring
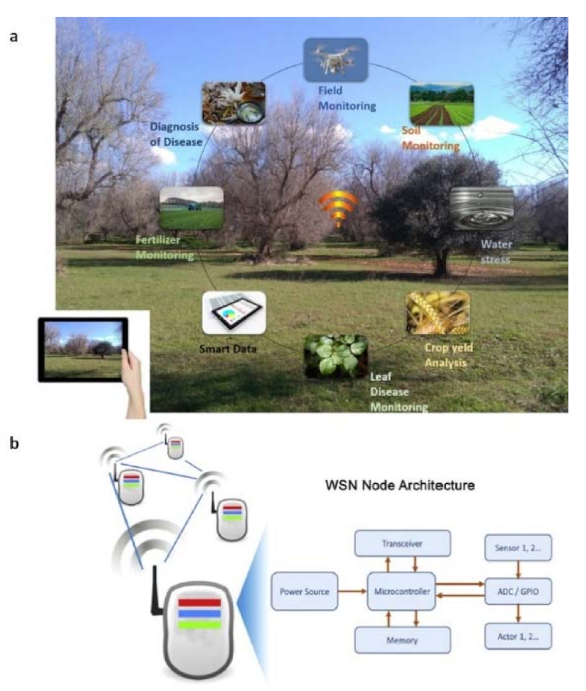
3.5. IoT and Remote Sensing Technologies
4. Discussion and Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Brasier, C.M. The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 2008, 57, 792–808. [Google Scholar] [CrossRef]
- Miller, S.A.; Beed, F.D.; Harmon, C.L. Plant disease diagnostic capabilities and networks. Annu. Rev. Phytopathol. 2009, 47, 15–38. [Google Scholar] [CrossRef]
- Strange, R.N.; Scott, P.R. Plant Disease: A Threat to Global Food Security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef]
- Oerke, E.C.; Dehne, H.W.; Schonbeck, F.; Weber, A. Crop Production and Crop Protection. Estimated Losses in Major Food and Cash Crops; Elsevier Science: Amsterdam, The Nederlands, 1994. [Google Scholar] [CrossRef]
- Oerke, E.C. Crop Losses to Pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Rybicki, E.P. A Top Ten list for economically important plant viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef]
- Scholthof, K.B.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. Available online: https://www.eppo.int/ACTIVITIES/plant_quarantine/A2_list (accessed on 7 October 2020).
- Saponari, M.; Boscia, D.; Nigro, F.; Martelli, G. Identification of Dna Sequences Related to Xylella fastidiosa in Oleander, Almond and Olive Trees Exhibiting Leaf Scorch Symptoms in Apulia (southern Italy). J. Plant Pathol. 2013, 95, 668. [Google Scholar]
- Regione Puglia. Puglia Region Bulletin Number 39 of 2015. Available online: http://burp.regione.puglia.it/bollettino-ufficiale?p_p_id=burpsearch_WAR_GestioneBurpportlet&p_p_lifecycle=0&p_p_state=normal&p_p_mode=view&p_p_col_id=column-1&p_p_col_count=1&_burpsearch_WAR_GestioneBurpportlet_jspPage=%2Fhtml%2Fburpsearch%2Fview.jsp&_burpsearch_WAR_GestioneBurpportlet_opz=dettagliosezione&_burpsearch_WAR_GestioneBurpportlet_anno=2015&_burpsearch_WAR_GestioneBurpportlet_burpId=15744&_burpsearch_WAR_GestioneBurpportlet_sezioneId=4574629 (accessed on 17 December 2020).
- Regione Puglia. Puglia Region Bulletin Number 157 of 2020. Available online: http://burp.regione.puglia.it/bollettino-ufficiale (accessed on 17 December 2020).
- Janse, J.; Obradovi, A. Xylella fastidiosa: Its biology, diagnosis, control and risks. J. Plant Pathol. 2010, 92, S35–S48. [Google Scholar]
- Martelli, G.P.; Boscia, D.; Porcelli, F.; Saponari, M. The olive quick decline syndrome in south-east Italy: A threatening phytosanitary emergency. Eur. J. Plant Pathol. 2016, 144, 235–243. [Google Scholar] [CrossRef]
- Sicard, A.; Zeilinger, A.R.; Vanhove, M.; Schartel, T.E.; Beal, D.J.; Daugherty, M.P.; Almeida, R.P.P. Xylella fastidiosa: Insights into an Emerging Plant Pathogen. Annu. Rev. Phytopathol. 2018, 56, 181–202. [Google Scholar] [CrossRef]
- Coker, T.L.R.; Rozsypálek, J.; Edwards, A.; Harwood, T.P.; Butfoy, L.; Buggs, R.J.A. Estimating mortality rates of European ash (Fraxinus excelsior) under the ash dieback (Hymenoscyphus fraxineus) epidemic. Plants People Planet 2019, 1, 48–58. [Google Scholar] [CrossRef]
- The Governing Body of the International Plant Protection Convention, Which Is in Charge of Establishing and Implementing Phytosanitary Standards Recognized by Governments around the World. Available online: https://www.ippc.int/en/ (accessed on 17 December 2020).
- Nezhad, A.S. Future of portable devices for plant pathogen diagnosis. Lab Chip 2014, 14, 2887–2904. [Google Scholar] [CrossRef]
- EUR-Lex. Regulation (EU) 2016/2031 of the European Parliament of the Council of 26 October 2016 on Protective Measures against Pests of Plants, Amending Regulations (EU) No 228/2013, (EU) No 652/2014 and (EU) No 1143/2014 of the European Parliament and of the Council and Repealing Council Directives 69/464/EEC, 74/647/EEC, 93/85/EEC, 98/57/EC, 2000/29/EC, 2006/91/EC and 2007/33/EC. Available online: http://data.europa.eu/eli/reg/2016/2031/oj (accessed on 7 October 2020).
- EUR-Lex. Regulation (EU) 2017/625 of the European Parliament and of the Council of 15 March 2017 on Official Controls and Other Official Activities Performed to Ensure the Application of Food and Feed Law, Rules on Animal Health and Welfare, Plant Health and Plant Protection Products, Amending Regulations (EC) No 999/2001, (EC) No 396/2005, (EC) No 1069/2009, (EC) No 1107/2009, (EU) No 1151/2012, (EU) No 652/2014, (EU) 2016/429 and (EU) 2016/2031 of the European Parliament and of the Council, Council Regulations (EC) No 1/2005 and (EC) No 1099/2009 and Council Directives 98/58/EC, 1999/74/EC, 2007/43/EC, 2008/119/EC and 2008/120/EC, and Repealing Regulations (EC) No 854/2004 and (EC) No 882/2004 of the European Parliament and of the Council, Council Directives 89/608/EEC, 89/662/EEC, 90/425/EEC, 91/496/EEC, 96/23/EC, 96/93/EC and 97/78/EC and Council Decision 92/438/EEC (Official Controls Regulation) Text with EEA Relevance. Available online: http://data.europa.eu/eli/reg/2017/625/oj (accessed on 7 October 2020).
- Vashist, S.K. Point-of-Care Diagnostics: Recent Advances and Trends. Biosensors 2017, 7, 62. [Google Scholar] [CrossRef]
- Kettler, H.; White, K.; Hawkes, S.J. Mapping the Landscape of Diagnostics for Sexually Transmitted Infections: Key Findings and Recommendations; World Health Organization: Geneva, Switzerland, 2004. [Google Scholar]
- Point of Care Diagnostics Market by Product (Glucose, Infectious Disease (Hepatitis C, Influenza, Respiratory), Coagulation), Platform (Microfluidics, Immunoassays), Mode (Prescription & OTC), End-User (Hospitals, Home Care)—Global Forecast to 2024. Available online: https://www.marketsandmarkets.com/Market-Reports/point-of-care-diagnostic-market-106829185.html (accessed on 7 October 2020).
- Pai, N.; Ghiasi, M.; Pai, M. Point-of-Care Diagnostic Testing in Global Health: What Is the Point? Microbe Mag. 2015, 10, 103–107. [Google Scholar] [CrossRef]
- Manessis, G.; Gelasakis, A.; Bossis, Y. The challenge of introducing Point of Care Diagnostics in Farm Animal Health Management. Biomed. J. Sci. Tech. Res. 2019, 14. [Google Scholar] [CrossRef]
- Lau, H.Y.; Botella, J.R. Advanced DNA-Based Point-of-Care Diagnostic Methods for Plant Diseases Detection. Front. Plant Sci. 2017, 8, 2016. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.; Foster, S.; Fraaije, B.; McCartney, H. Plant pathogen diagnostics: Immunological and nucleic acid-based approaches. Ann. Appl. Biol. 2005, 145, 1–16. [Google Scholar] [CrossRef]
- Alvarez, A.M. Integrated approaches for detection of plant pathogenic bacteria and diagnosis of bacterial diseases. Annu. Rev. Phytopathol. 2004, 42, 339–366. [Google Scholar] [CrossRef] [PubMed]
- López, M.; Bertolini, E.; Caruso, P.; Penyalver, R.; Marco-Noales, E.; Gorris, M.; Morente, C.; Salcedo, C.; Cambra, M.; Llop, P. Advantages of an integrated approach for diagnosis of quarantine pathogenic bacteria in plant material. Phytopathol. Pol. 2005, 35, 49–56. [Google Scholar]
- López, M.M.; Bertolini, E.; Olmos, A.; Caruso, P.; Gorris, M.T.; Llop, P.; Penyalver, R.; Cambra, M. Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2003, 6, 233–243. [Google Scholar] [CrossRef]
- Fang, Y.; Ramasamy, R.P. Current and Prospective Methods for Plant Disease Detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Dincer, C.; Bruch, R.; Kling, A.; Dittrich, P.S.; Urban, G.A. Multiplexed Point-of-Care Testing—xPOCT. Trends Biotechnol. 2017, 35, 728–742. [Google Scholar] [CrossRef]
- Zarei, M. Advances in point-of-care technologies for molecular diagnostics. Biosens. Bioelectron. 2017, 98, 494–506. [Google Scholar] [CrossRef]
- Zarei, M. Portable biosensing devices for point-of-care diagnostics: Recent developments and applications. Trac. Trends Anal. Chem. 2017, 91, 26–41. [Google Scholar] [CrossRef]
- Huang, X.; Xu, J.; Ji, H.-F.; Li, G.; Chen, H. Quartz crystal microbalance based biosensor for rapid and sensitive detection of maize chlorotic mottle virus. Anal. Methods 2014, 6, 4530–4536. [Google Scholar] [CrossRef]
- Sauerbrey, G. Verwendung von Schwingquarzen zur Wägung dünner Schichten und zur Mikrowägung. Z. Phys. 1959, 155, 206–222. [Google Scholar] [CrossRef]
- Montagut, Y.; Narbon, J.G.; Jimenez, Y.; March, C.; Montoya, A.; Arnau, A. QCM Technology in Biosensors. Biosens. Emerg. Mater. Appl. 2011, 153–178. [Google Scholar] [CrossRef]
- Lin, H.Y.; Huang, C.H.; Lu, S.H.; Kuo, I.T.; Chau, L.K. Direct detection of orchid viruses using nanorod-based fiber optic particle plasmon resonance immunosensor. Biosens. Bioelectron. 2014, 51, 371–378. [Google Scholar] [CrossRef]
- Pan, T.-T.; Sun, D.-W.; Pu, H.; Wei, Q. Simple Approach for the Rapid Detection of Alternariol in Pear Fruit by Surface-Enhanced Raman Scattering with Pyridine-Modified Silver Nanoparticles. J. Agric. Food Chem. 2018, 66, 2180–2187. [Google Scholar] [CrossRef]
- Katz, E.; Willner, I. Probing Biomolecular Interactions at Conductive and Semiconductive Surfaces by Impedance Spectroscopy: Routes to Impedimetric Immunosensors, DNA-Sensors, and Enzyme Biosensors. Electroanalysis 2003, 15, 913–947. [Google Scholar] [CrossRef]
- Jócsák, I.; Végvári, G.; Vozáry, E. Electrical impedance measurement on plants: A review with some insights to other fields. Theor. Exp. Plant Physiol. 2019, 31, 359–375. [Google Scholar] [CrossRef]
- Khater, M.; de la Escosura-Muñiz, A.; Quesada-González, D.; Merkoçi, A. Electrochemical detection of plant virus using gold nanoparticle-modified electrodes. Anal. Chim. Acta 2019, 1046, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Jarocka, U.; Wąsowicz, M.; Radecka, H.; Malinowski, T.; Michalczuk, L.; Radecki, J. Impedimetric Immunosensor for Detection of Plum Pox Virus in Plant Extracts. Electroanalysis 2011, 23, 2197–2204. [Google Scholar] [CrossRef]
- Jarocka, U.; Radecka, H.; Malinowski, T.; Michalczuk, L.; Radecki, J. Detection of Prunus Necrotic Ringspot Virus in Plant Extracts with Impedimetric Immunosensor based on Glassy Carbon Electrode. Electroanalysis 2013, 25. [Google Scholar] [CrossRef]
- Chiriacò, M.S.; Luvisi, A.; Primiceri, E.; Sabella, E.; De Bellis, L.; Maruccio, G. Development of a lab-on-a-chip method for rapid assay of Xylella fastidiosa subsp. pauca strain CoDiRO. Sci. Rep. 2018, 8, 7376. [Google Scholar] [CrossRef]
- Cebula, Z.; Żołędowska, S. Detection of the Plant Pathogen Pseudomonas Syringae pv. Lachrymans on Antibody-Modified Gold Electrodes by Electrochemical Impedance Spectroscopy. Sensors 2019, 19, 5411. [Google Scholar] [CrossRef] [PubMed]
- EUR-Lex. Council Directive 2008/90/EC of 29 September 2008 on the Marketing of Fruit Plant Propagating Material and Fruit Plants Intended for Fruit Production (Recast Version). Available online: http://data.europa.eu/eli/dir/2008/90/oj (accessed on 7 October 2020).
- EUR-Lex. Council Directive 2002/11/EC of 14 February 2002 Amending Directive 68/193/EEC on the Marketing of Material for the Vegetative Propagation of the Vine and Repealing Directive 74/649/EEC. Available online: http://data.europa.eu/eli/dir/2002/11/oj (accessed on 7 October 2020).
- EUR-Lex. Commission Directive 2005/43/EC of 23 June 2005 Amending the Annexes to Council Directive 68/193/EEC on the Marketing of Material for the Vegetative Propagation of the Vine. Available online: http://data.europa.eu/eli/dir/2005/43/oj (accessed on 7 October 2020).
- Scala, A.; Allmann, S.; Mirabella, R.; Haring, M.; Schuurink, R. Green Leaf Volatiles: A Plant’s Multifunctional Weapon against Herbivores and Pathogens. Int. J. Mol. Sci. 2013, 14, 17781–17811. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, I.; Halitschke, R.; Paschold, A.; Dahl, C.; Preston, C. Volatile Signaling in Plant-Plant Interactions: “Talking Trees” in the Genomics Era. Science 2006, 311, 812–815. [Google Scholar] [CrossRef] [PubMed]
- Cellini, A.; Biondi, E.; Blasioli, S.; Rocchi, L.; Farneti, B.; Braschi, I.; Savioli, S.; Rodriguez-Estrada, M.; Biasioli, F.; Spinelli, F. Early detection of bacterial diseases in apple plants by analysis of volatile organic compounds profiles and use of electronic nose. Ann. Appl. Biol. 2016, 168. [Google Scholar] [CrossRef]
- De Lacy Costello, B.; Evans, P.; Ewen, R.; Gunson, H.; Jones, P.; Ratcliffe, N.; Spencer-Phillips, P. Gas chromatography–mass spectrometry analyses of volatile organic compounds from potato tubers inoculated with Phytophthora infestans or Fusarium coeruleum. Plant Pathol. 2001, 50, 489–496. [Google Scholar] [CrossRef]
- Ewen, R.J.; Jones, P.R.H.; Ratcliffe, N.M.; Spencer-Phillips, P.T.N. Identification by gas chromatography-mass spectrometry of the volatile organic compounds emitted from the wood-rotting fungi Serpula lacrymans and Coniophora puteana, and from Pinus sylvestris timber. Mycol. Res. 2004, 108, 806–814. [Google Scholar] [CrossRef]
- Spadafora, N.; Paramithiotis, S.; Drosinos, E.; Cammarisano, L.; Rogers, H.; Muller, C.T. Detection of Listeria monocytogenes in cut melon fruit using analysis of volatile organic compounds. Food Microbiol. 2016, 54, 52–59. [Google Scholar] [CrossRef]
- Sharma, R.; Zhou, M.; Hunter, M.D.; Fan, X. Rapid In Situ Analysis of Plant Emission for Disease Diagnosis Using a Portable Gas Chromatography Device. J. Agric. Food Chem. 2019, 67, 7530–7537. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Sanati Nezhad, A. Microfluidic platforms for plant cells studies. Lab Chip 2014, 14, 3262–3274. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Wilson, A. Applications of Electronic-Nose Technologies for Noninvasive Early Detection of Plant, Animal and Human Diseases. Chemosensors 2018, 6, 45. [Google Scholar] [CrossRef]
- Persaud, K.; Dodd, G. Analysis of discrimination mechanisms in the mammalian olfactory system using a model nose. Nature 1982, 299, 352–355. [Google Scholar] [CrossRef]
- Wilson, A.D.; Baietto, M. Applications and advances in electronic-nose technologies. Sensors 2009, 9, 5099–5148. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Diverse applications of electronic-nose technologies in agriculture and forestry. Sensors 2013, 13, 2295–2348. [Google Scholar] [CrossRef] [PubMed]
- Laothawornkitkul, J.; Moore, J.P.; Taylor, J.E.; Possell, M.; Gibson, T.D.; Hewitt, C.N.; Paul, N.D. Discrimination of Plant Volatile Signatures by an Electronic Nose: A Potential Technology for Plant Pest and Disease Monitoring. Environ. Sci. Technol. 2008, 42, 8433–8439. [Google Scholar] [CrossRef]
- Ampuero, S.; Bosset, J.O. The electronic nose applied to dairy products: A review. Sens. Actuators B Chem. 2003, 94, 1–12. [Google Scholar] [CrossRef]
- Peris, M.; Escuder-Gilabert, L. A 21st century technique for food control: Electronic noses. Anal. Chim. Acta 2009, 638, 1–15. [Google Scholar] [CrossRef]
- Kalman, E.-L.; Löfvendahl, A.; Winquist, F.; Lundström, I. Classification of complex gas mixtures from automotive leather using an electronic nose. Anal. Chim. Acta 2000, 403, 31–38. [Google Scholar] [CrossRef]
- Cui, S.; Ling, P.; Zhu, H.; Keener, H.M. Plant Pest Detection Using an Artificial Nose System: A Review. Sensors 2018, 18, 378. [Google Scholar] [CrossRef]
- Chalupowicz, D.; Veltman, B.; Droby, S.; Eltzov, E. Evaluating the use of biosensors for monitoring of Penicillium digitatum infection in citrus fruit. Sens. Actuators B Chem. 2020, 311, 127896. [Google Scholar] [CrossRef]
- Wang, H.; Ramnani, P.; Pham, T.; Villarreal, C.C.; Yu, X.; Liu, G.; Mulchandani, A. Gas Biosensor Arrays Based on Single-Stranded DNA-Functionalized Single-Walled Carbon Nanotubes for the Detection of Volatile Organic Compound Biomarkers Released by Huanglongbing Disease-Infected Citrus Trees. Sensors 2019, 19, 4795. [Google Scholar] [CrossRef]
- Staii, C.; Johnson, A.T.; Chen, M.; Gelperin, A. DNA-Decorated Carbon Nanotubes for Chemical Sensing. Nano Lett. 2005, 5, 1774–1778. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Paul, R.; Ba Tis, T.; Saville, A.C.; Hansel, J.C.; Yu, T.; Ristaino, J.B.; Wei, Q. Non-invasive plant disease diagnostics enabled by smartphone-based fingerprinting of leaf volatiles. Nat. Plants 2019, 5, 856–866. [Google Scholar] [CrossRef]
- McDonald, J.C.; Duffy, D.C.; Anderson, J.R.; Chiu, D.T.; Wu, H.; Schueller, O.J.; Whitesides, G.M. Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 2000, 21, 27–40. [Google Scholar] [CrossRef]
- Ducrée, J. Special Issue: Microfluidic Lab-on-a-Chip Platforms for High-Performance Diagnostics. Diagnostics 2012, 2, 1. [Google Scholar] [CrossRef]
- Foudeh, A.M.; Fatanat Didar, T.; Veres, T.; Tabrizian, M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 2012, 12, 3249–3266. [Google Scholar] [CrossRef]
- Khandurina, J.; Guttman, A. Bioanalysis in microfluidic devices. J. Chromatogr. A 2002, 943, 159–183. [Google Scholar] [CrossRef]
- Fang, X.; Chen, H.; Yu, S.; Jiang, X.; Kong, J. Predicting viruses accurately by a multiplex microfluidic loop-mediated isothermal amplification chip. Anal. Chem. 2011, 83, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Huang, J.-G.; Chuang, T.-L.; Sheu, J.-C.; Chuang, Y.-K.; Holl, M.; Meldrum, D.R.; Lee, C.-N.; Lin, C.-W. Compact optical diagnostic device for isothermal nucleic acids amplification. Sens Actuators B Chem 2008, 133, 493–501. [Google Scholar] [CrossRef]
- Dharmasiri, U.; Witek, M.A.; Adams, A.A.; Osiri, J.K.; Hupert, M.L.; Bianchi, T.S.; Roelke, D.L.; Soper, S.A. Enrichment and detection of Escherichia coli O157:H7 from water samples using an antibody modified microfluidic chip. Anal. Chem. 2010, 82, 2844–2849. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Tokunaga, Y.; Goto, S.; Fujii, Y.; Banno, F.; Edagawa, A. Rapid on-site monitoring of Legionella pneumophila in cooling tower water using a portable microfluidic system. Sci. Rep. 2017, 7, 3092. [Google Scholar] [CrossRef] [PubMed]
- Tourlousse, D.M.; Ahmad, F.; Stedtfeld, R.D.; Seyrig, G.; Tiedje, J.M.; Hashsham, S.A. A polymer microfluidic chip for quantitative detection of multiple water- and foodborne pathogens using real-time fluorogenic loop-mediated isothermal amplification. Biomed. Microdevices 2012, 14, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Trinh, K.T.L.; Zhang, H.; Kang, D.-J.; Kahng, S.-H.; Tall, B.D.; Lee, N.Y. Fabrication of Polymerase Chain Reaction Plastic Lab-on-a-Chip Device for Rapid Molecular Diagnoses. Int. Neurourol. J. 2016, 20, S38–S48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Bi, H.; Liu, B. Detection of Pathogenic Microorganisms by Microfluidics Based Analytical Methods. Anal. Chem. 2018, 90, 5512–5520. [Google Scholar] [CrossRef] [PubMed]
- Julich, S.; Riedel, M.; Kielpinski, M.; Urban, M.; Kretschmer, R.; Wagner, S.; Fritzsche, W.; Henkel, T.; Möller, R.; Werres, S. Development of a lab-on-a-chip device for diagnosis of plant pathogens. Biosens. Bioelectron. 2011, 26, 4070–4075. [Google Scholar] [CrossRef] [PubMed]
- Schwenkbier, L.; Pollok, S.; König, S.; Urban, M.; Werres, S.; Cialla-May, D.; Weber, K.; Popp, J. Towards on-site testing of Phytophthora species. Anal. Methods 2015, 7, 211–217. [Google Scholar] [CrossRef]
- Chang, W.-H.; Yang, S.-Y.; Lin, C.-L.; Wang, C.-H.; Li, P.-C.; Chen, T.-Y.; Jan, F.-J.; Lee, G.-B. Detection of viruses directly from the fresh leaves of a Phalaenopsis orchid using a microfluidic system. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 1274–1282. [Google Scholar] [CrossRef]
- Lin, C.-L.; Chang, W.-H.; Wang, C.-H.; Lee, C.-H.; Chen, T.-Y.; Jan, F.-J.; Lee, G.-B. A microfluidic system integrated with buried optical fibers for detection of Phalaenopsis orchid pathogens. Biosens. Bioelectron. 2015, 63, 572–579. [Google Scholar] [CrossRef]
- Qu, X.; Li, M.; Zhang, H.; Lin, C.; Wang, F.; Xiao, M.; Zhou, Y.; Shi, J.; Aldalbahi, A.; Pei, H.; et al. Real-Time Continuous Identification of Greenhouse Plant Pathogens Based on Recyclable Microfluidic Bioassay System. ACS Appl. Mater. Interfaces 2017, 9, 31568–31575. [Google Scholar] [CrossRef]
- De La Fuente, L.; Montanes, E.; Meng, Y.; Li, Y.; Burr, T.J.; Hoch, H.C.; Wu, M. Assessing adhesion forces of type I and type IV pili of Xylella fastidiosa bacteria by use of a microfluidic flow chamber. Appl. Environ. Microbiol. 2007, 73, 2690–2696. [Google Scholar] [CrossRef]
- Meng, Y.; Li, Y.; Galvani, C.D.; Hao, G.; Turner, J.N.; Burr, T.J.; Hoch, H.C. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J. Bacteriol. 2005, 187, 5560–5567. [Google Scholar] [CrossRef]
- Neethirajan, S.; Kobayashi, I.; Nakajima, M.; Wu, D.; Nandagopal, S.; Lin, F. Microfluidics for food, agriculture and biosystems industries. Lab Chip 2011, 11, 1574–1586. [Google Scholar] [CrossRef]
- Nezhad, A.S.; Naghavi, M.; Packirisamy, M.; Bhat, R.; Geitmann, A. Quantification of the Young's modulus of the primary plant cell wall using Bending-Lab-On-Chip (BLOC). Lab Chip 2013, 13, 2599–2608. [Google Scholar] [CrossRef]
- Grossmann, G.; Guo, W.J.; Ehrhardt, D.W.; Frommer, W.B.; Sit, R.V.; Quake, S.R.; Meier, M. The RootChip: An integrated microfluidic chip for plant science. Plant Cell 2011, 23, 4234–4240. [Google Scholar] [CrossRef]
- Grossmann, G.; Meier, M.; Cartwright, H.N.; Sosso, D.; Quake, S.R.; Ehrhardt, D.W.; Frommer, W.B. Time-lapse fluorescence imaging of Arabidopsis root growth with rapid manipulation of the root environment using the RootChip. J. Vis. Exp. 2012, 65, e4290. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Körner, O.; Challa, H. Process-based humidity control regime for greenhouse crops. Comput. Electron. Agric. 2003, 39, 173–192. [Google Scholar] [CrossRef]
- Mortensen, L. Effects of air humidity on growth, flowering, keeping quality and water relations of four short-day greenhouse species. Sci. Hortic. 2000, 86, 299–310. [Google Scholar] [CrossRef]
- Sánchez-Blanco, M.J.; Ferrández, T.; Navarro, A.; Bañon, S.; Alarcón, J.J. Effects of irrigation and air humidity preconditioning on water relations, growth and survival of Rosmarinus officinalis plants during and after transplanting. J. Plant Physiol. 2004, 161, 1133–1142. [Google Scholar] [CrossRef]
- Sunkar, R. Plant Stress Tolerance. Methods Protoc. 2010, 639, 401. [Google Scholar] [CrossRef]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Lee, K.; Park, J.; Lee, M.S.; Kim, J.; Hyun, B.G.; Kang, D.J.; Na, K.; Lee, C.Y.; Bien, F.; Park, J.U. In-situ synthesis of carbon nanotube-graphite electronic devices and their integrations onto surfaces of live plants and insects. Nano Lett. 2014, 14, 2647–2654. [Google Scholar] [CrossRef]
- Koman, V.B.; Lew, T.T.S.; Wong, M.H.; Kwak, S.Y.; Giraldo, J.P.; Strano, M.S. Persistent drought monitoring using a microfluidic-printed electro-mechanical sensor of stomata in planta. Lab Chip 2017, 17, 4015–4024. [Google Scholar] [CrossRef]
- Tang, W.; Yan, T.; Wang, F.; Yang, J.; Wu, J.; Wang, J.; Yue, T.; Li, Z. Rapid fabrication of wearable carbon nanotube/graphite strain sensor for real-time monitoring of plant growth. Carbon 2019, 147, 295–302. [Google Scholar] [CrossRef]
- Nassar, J.M.; Khan, S.M.; Villalva, D.R.; Nour, M.M.; Almuslem, A.S.; Hussain, M.M. Compliant plant wearables for localized microclimate and plant growth monitoring. NPJ Flex. Electron. 2018, 2, 24. [Google Scholar] [CrossRef]
- Coppedè, N.; Janni, M.; Bettelli, M.; Maida, C.L.; Gentile, F.; Villani, M.; Ruotolo, R.; Iannotta, S.; Marmiroli, N.; Marmiroli, M.; et al. An in vivo biosensing, biomimetic electrochemical transistor with applications in plant science and precision farming. Sci. Rep. 2017, 7, 16195. [Google Scholar] [CrossRef] [PubMed]
- Oren, S.; Ceylan, H.; Schnable, P.; Dong, L. Wearable Electronics: High-Resolution Patterning and Transferring of Graphene-Based Nanomaterials onto Tape toward Roll-to-Roll Production of Tape-Based Wearable Sensors (Adv. Mater. Technol. 12/2017). Adv. Mater. Technol. 2017, 2, 1770055. [Google Scholar] [CrossRef]
- Maksimovic, M.; Omanovic-Miklicanin, E. Green Internet of Things and Green nanotechnology role in realizing smart and sustainable agriculture. In Proceedings of the VIII International Scientific Agriculture Symposium “AGROSYM 2017”, Jahorina, Bosnia, 5–8 October 2017; Kovacevic, D., Ed.; University of East Sarajevo: Sarajevo, Bosnia, 2017; pp. 2290–2295. [Google Scholar]
- Bastiaanssen, W.G.M.; Molden, D.J.; Makin, I.W. Remote sensing for irrigated agriculture: Examples from research and possible applications. Agric. Water Manag. 2000, 46, 137–155. [Google Scholar] [CrossRef]
- Hashem, I.A.T.; Yaqoob, I.; Anuar, N.B.; Mokhtar, S.; Gani, A.; Ullah Khan, S. The rise of “big data” on cloud computing: Review and open research issues. Inf. Syst. 2015, 47, 98–115. [Google Scholar] [CrossRef]
- Weber, R.; Romana, W. Internet of Things. Legal Perspectives; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Ampatzidis, Y.; De Bellis, L.; Luvisi, A. iPathology: Robotic Applications and Management of Plants and Plant Diseases. Sustainability 2017, 9, 1010. [Google Scholar] [CrossRef]
- Mulla, D.J. Twenty five years of remote sensing in precision agriculture: Key advances and remaining knowledge gaps. Biosyst. Eng. 2013, 114, 358–371. [Google Scholar] [CrossRef]
- Meroni, M.R.M.; Colombo, R. Characterization of leaf physiology using reflectance and fluorescence hyperspectral measurements. In Optical Observation of Vegetation Properties and Characteristics; Maselli, F.M.M., Brivio, P.A., Eds.; Research Signpost: Trivandrum, India, 2010. [Google Scholar]
- West, J.S.; Bravo, C.; Oberti, R.; Lemaire, D.; Moshou, D.; McCartney, H.A. The potential of optical canopy measurement for targeted control of field crop diseases. Annu. Rev. Phytopathol. 2003, 41, 593–614. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zeng, A.; Zhu, T.; Fang, S.; Gong, Y.; Tao, Y.; Zhou, Y.; Liu, K. Using remotely sensed spectral reflectance to indicate leaf photosynthetic efficiency derived from active fluorescence measurements. J. Appl. Remote Sens. 2017, 11, 026034. [Google Scholar] [CrossRef]
- SenseFly-The Professional’s Mapping Drone. Available online: https://www.sensefly.com/ (accessed on 7 October 2020).
- Omasa, K.; Oki, K.; Suhama, T. 5.2 Remote Sensing from Satellites and Aircraft; ASABE: St. Joseph, MI, USA, 2006. [Google Scholar] [CrossRef]
- Rudd, J.D.; Roberson, G.T.; Classen, J.J. Application of satellite, unmanned aircraft system, and ground-based sensor data for precision agriculture: A review. In Proceedings of the 2017 ASABE Annual International Meeting, St. Joseph, MI, USA, 16–19 July 2017; p. 1. [Google Scholar]
- Bendig, J.; Bolten, A.; Bareth, G. Introducing a low-cost mini-UAV for thermal-and multispectral-imaging. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, XXXIX–B1, 345–349. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Huete, A. Remote Sensing for Environmental Monitoring; Academic Press: New York, NY, USA, 2004; pp. 183–206. [Google Scholar] [CrossRef]
- Ashton, K. That “Internet of Things” Thing. RFID J. 2009, 22, 97–114. [Google Scholar]
- Luvisi, A.; Ampatzidis, Y.; De Bellis, L. Plant Pathology and Information Technology: Opportunity for Management of Disease Outbreak and Applications in Regulation Frameworks. Sustainability 2016, 8, 831. [Google Scholar] [CrossRef]
- Publications Office of the European Union. Vision and Challenges for Realising the Internet of Things; Publications Office of the European Union: Luxembourg, 2010. [Google Scholar] [CrossRef]
- Tzounis, A.; Katsoulas, N.; Bartzanas, T.; Kittas, C. Internet of Things in agriculture, recent advances and future challenges. Biosyst. Eng. 2017, 164, 31–48. [Google Scholar] [CrossRef]
- Atzori, L.; Iera, A.; Morabito, G. The Internet of Things: A survey. Comput. Netw. 2010, 54, 2787–2805. [Google Scholar] [CrossRef]
- Botta, A.; Donato, W.d.; Persico, V.; Pescapé, A. Integration of Cloud computing and Internet of Things. Future Gener. Comput. Syst. 2016, 56, 684–700. [Google Scholar] [CrossRef]
- Gubbi, J.; Buyya, R.; Marusic, S.; Palaniswami, M. Internet of Things (IoT): A vision, architectural elements, and future directions. Future Gener. Comput. Syst. 2013, 29, 1645–1660. [Google Scholar] [CrossRef]
- Farooq, M.S.; Riaz, S.; Abid, A.; Umer, T.; Zikria, Y.B. Role of IoT Technology in Agriculture: A Systematic Literature Review. Electronics 2020, 9, 319. [Google Scholar] [CrossRef]
- Ray, P.P. Internet of things for smart agriculture: Technologies, practices and future direction. J. Ambient Intell. Smart Environ. 2017, 9, 395–420. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Z. Application of big data technology in agricultural Internet of Things. Int. J. Distrib. Sens. Netw. 2019, 15, 155014771988161. [Google Scholar] [CrossRef]
- Liqiang, Z.; Shouyi, Y.; Leibo, L.; Zhen, Z.; Shaojun, W. A Crop Monitoring System Based on Wireless Sensor Network. Procedia Environ. Sci. 2011, 11, 558–565. [Google Scholar] [CrossRef]
- Jing, Y.; Yuzhi, Z.; Dan, D.; Xiao, W.; Ping, Y.; Ling-fang, C.; Yue-fang, S.; Ze-tao, L. An early warning system of diseases and pests for blueberry based on WSN. In Proceedings of the 2017 36th Chinese Control Conference (CCC), Dalian, China, 26–28 July 2017; pp. 8885–8889. [Google Scholar]
- Song, Y.; Ma, J.; Zhang, X.; Feng, Y. Design of Wireless Sensor Network-Based Greenhouse Environment Monitoring and Automatic Control System. J. Netw. 2012, 7, 838–844. [Google Scholar] [CrossRef]
- Luvisi, A.; Panattoni, A.; Bandinelli, R.; Rinaldelli, E.; Pagano, M.; Triolo, E. Implanting RFIDs into Prunus to facilitate electronic identification in support of sanitary certification. Biosyst. Eng. 2011, 109, 167–173. [Google Scholar] [CrossRef]
- Pagano, M.; Bandinelli, R.; Rinaldelli, E.; Panattoni, A.; Triolo, E.; Luvisi, A. RFID technology for clonal selection purposes. Adv. Hortic. Sci. 2010, 24, 282–284. [Google Scholar]
- Luvisi, A.; Panattoni, A.; Bandinelli, E.; Rinaldelli, M.P.; Triolo, E. Propagative material of grapevine: RFID technology for supporting traceability of “basic” and “certified” material along the wine production chain. Adv. Hortic. Sci. 2012, 26, 39–43. [Google Scholar] [CrossRef]
- Rupanagudi, S.R.; Ranjani, B.S.; Nagaraj, P.; Bhat, V.G.; Thippeswamy, G. A novel cloud computing based smart farming system for early detection of borer insects in tomatoes. In Proceedings of the 2015 International Conference on Communication, Information & Computing Technology (ICCICT), Mumbai, India, 15–17 January 2015; pp. 1–6. [Google Scholar]
- Pérez-Expósito, J.P.; Fernández-Caramés, T.M.; Fraga-Lamas, P.; Castedo, L. VineSens: An Eco-Smart Decision-Support Viticulture System. Sensors 2017, 17, 465. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, F.; Bratanov, D. A Novel Methodology for Improving Plant Pest Surveillance in Vineyards and Crops Using UAV-Based Hyperspectral and Spatial Data. Sensors 2018, 18, 260. [Google Scholar] [CrossRef] [PubMed]
- Arable—Decision Agriculture. Available online: https://www.arable.com/ (accessed on 7 October 2020).
- Semios. Available online: https://semios.com/ (accessed on 7 October 2020).
- Poblete, T.; Camino, C.; Beck, P.S.A.; Hornero, A.; Kattenborn, T.; Saponari, M.; Boscia, D.; Navas-Cortes, J.A.; Zarco-Tejada, P.J. Detection of Xylella fastidiosa infection symptoms with airborne multispectral and thermal imagery: Assessing bandset reduction performance from hyperspectral analysis. ISPRS J. Photogramm. Remote Sens. 2020, 162, 27–40. [Google Scholar] [CrossRef]
- Hornero, A.; Hernandez Clemente, R.; North, P.R.J.; Beck, P.S.A.; Boscia, D.; Navas Cortés, J.; Zarco-Tejada, P. Monitoring the incidence of Xylella fastidiosa infection in olive orchards using ground-based evaluations, airborne imaging spectroscopy and Sentinel-2 time series through 3-D radiative transfer modelling. Remote Sens. Environ. 2020, 236, 111480. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, Y.-H.; Wu, Y.-Z.; Yue, Y. Agricultural Pests Tracking and Identification in Video Surveillance Based on Deep Learning. In International Conference on Intelligent Computing; Springer: Cham, Switzerland, 2017; pp. 58–70. [Google Scholar]
- Polo, J.; Hornero, G.; Duijneveld, C.; García, A.; Casas, O. Design of a low-cost Wireless Sensor Network with UAV mobile node for agricultural applications. Comput. Electron. Agric. 2015, 119, 19–32. [Google Scholar] [CrossRef]
- Vázquez-Arellano, M.; Griepentrog, H.W.; Reiser, D.; Paraforos, D.S. 3-D Imaging Systems for Agricultural Applications—A Review. Sensors 2016, 16, 618. [Google Scholar] [CrossRef]
- Kamilaris, A.; Andreas, K.; Francesc, X.P.-B. A review on the practice of big data analysis in agriculture. Comput. Electron. Agric. 2017, 143, 23–37. [Google Scholar] [CrossRef]
- Balasubramaniam, S.; Kangasharju, J. Realizing the Internet of Nano Things: Challenges, Solutions, and Applications. Computer 2013, 46, 62–68. [Google Scholar] [CrossRef]
- Cruz Alvarado, M.A.; Bazán, P. Understanding the Internet of Nano Things: Overview, trends, and challenges. E-Cienc. De La Inf. 2019, 9, 152–182. [Google Scholar] [CrossRef]
- Priye, A.; Wong, S.; Bi, Y.; Carpio, M.; Chang, J.; Coen, M.; Cope, D.; Harris, J.; Johnson, J.; Keller, A.; et al. Lab-on-a-Drone: Toward Pinpoint Deployment of Smartphone-Enabled Nucleic Acid-Based Diagnostics for Mobile Health Care. Anal. Chem. 2016, 88, 4651–4660. [Google Scholar] [CrossRef]
- Mendes, J.; Pinho, T.M.; Neves Dos Santos, F.; Sousa, J.; Peres, E.; Cunha, J.; Cunha, M.; Morais, R. Smartphone Applications Targeting Precision Agriculture Practices—A Systematic Review. Agronomy 2020, 10, 855. [Google Scholar] [CrossRef]
- Pongnumkul, S.; Chaovalit, P.; Surasvadi, N. Applications of Smartphone-Based Sensors in Agriculture: A Systematic Review of Research. J. Sens. 2015, 2015, 195308. [Google Scholar] [CrossRef]
- Heeb, L.; Jenner, E.; Cock, M.J.W. Climate-smart pest management: Building resilience of farms and landscapes to changing pest threats. J. Pest Sci. 2019, 92, 951–969. [Google Scholar] [CrossRef]







| Techniques | Limit of Detection (CFU/mL) | Advantages | Limitations |
|---|---|---|---|
| PCR | 103–104 | Mature and common technology, portable, easy to operate | Effectiveness is subjected to DNA extraction, inhibitors, polymerase activity, concentration of PCR buffer, and deoxynucleoside triphosphate |
| FISH | 103 | High sensitivity | Autofluorescence, photobleaching |
| ELISA | 105–106 | Low cost, visual color change can be used for detection | Low sensitivity for bacteria |
| IF | 103 | High sensitivity, target distribution can be visualized | Photobleaching |
| FCM | 104 | Simultaneous measurement of several parameters, rapid detection | High cost, overwhelming unnecessary information |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buja, I.; Sabella, E.; Monteduro, A.G.; Chiriacò, M.S.; De Bellis, L.; Luvisi, A.; Maruccio, G. Advances in Plant Disease Detection and Monitoring: From Traditional Assays to In-Field Diagnostics. Sensors 2021, 21, 2129. https://doi.org/10.3390/s21062129
Buja I, Sabella E, Monteduro AG, Chiriacò MS, De Bellis L, Luvisi A, Maruccio G. Advances in Plant Disease Detection and Monitoring: From Traditional Assays to In-Field Diagnostics. Sensors. 2021; 21(6):2129. https://doi.org/10.3390/s21062129
Chicago/Turabian StyleBuja, Ilaria, Erika Sabella, Anna Grazia Monteduro, Maria Serena Chiriacò, Luigi De Bellis, Andrea Luvisi, and Giuseppe Maruccio. 2021. "Advances in Plant Disease Detection and Monitoring: From Traditional Assays to In-Field Diagnostics" Sensors 21, no. 6: 2129. https://doi.org/10.3390/s21062129
APA StyleBuja, I., Sabella, E., Monteduro, A. G., Chiriacò, M. S., De Bellis, L., Luvisi, A., & Maruccio, G. (2021). Advances in Plant Disease Detection and Monitoring: From Traditional Assays to In-Field Diagnostics. Sensors, 21(6), 2129. https://doi.org/10.3390/s21062129












