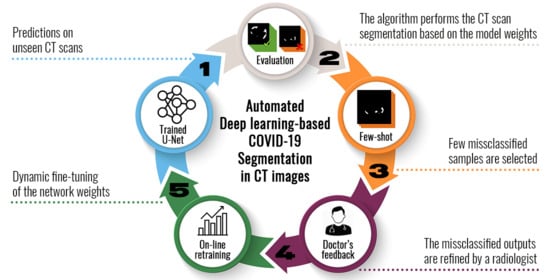
A Few-Shot U-Net Deep Learning Model for COVID-19 Infected Area Segmentation in CT Images
Abstract
Share and Cite
Voulodimos, A.; Protopapadakis, E.; Katsamenis, I.; Doulamis, A.; Doulamis, N. A Few-Shot U-Net Deep Learning Model for COVID-19 Infected Area Segmentation in CT Images. Sensors 2021, 21, 2215. https://doi.org/10.3390/s21062215
Voulodimos A, Protopapadakis E, Katsamenis I, Doulamis A, Doulamis N. A Few-Shot U-Net Deep Learning Model for COVID-19 Infected Area Segmentation in CT Images. Sensors. 2021; 21(6):2215. https://doi.org/10.3390/s21062215
Chicago/Turabian StyleVoulodimos, Athanasios, Eftychios Protopapadakis, Iason Katsamenis, Anastasios Doulamis, and Nikolaos Doulamis. 2021. "A Few-Shot U-Net Deep Learning Model for COVID-19 Infected Area Segmentation in CT Images" Sensors 21, no. 6: 2215. https://doi.org/10.3390/s21062215
APA StyleVoulodimos, A., Protopapadakis, E., Katsamenis, I., Doulamis, A., & Doulamis, N. (2021). A Few-Shot U-Net Deep Learning Model for COVID-19 Infected Area Segmentation in CT Images. Sensors, 21(6), 2215. https://doi.org/10.3390/s21062215