CovidArray: A Microarray-Based Assay with High Sensitivity for the Detection of Sars-Cov-2 in Nasopharyngeal Swabs
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Samples Collection
2.3. RNA Extraction and RT-qPCR
2.4. RNA Extraction for CovidArray Analysis
2.5. Reverse Transcription
2.6. PCR Conditions for Microarray
2.7. Microarray Preparation
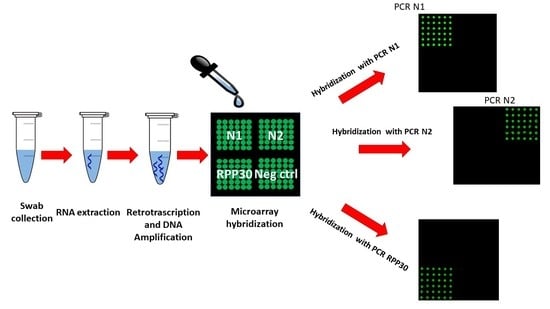
2.8. Microarray Hybridization and Detection
2.9. Droplet Digital PCR (ddPCR)
2.10. 2019-CoV Plasmid Controls
3. Results and Discussion
3.1. Detection of SARS-CoV-2 Nucleic Acid from Clinical Nasopharyngeal Swab Samples with CovidArray
3.2. Detection Limit of CovidArray
3.3. Multiplex Capability of the CovidArray
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar]
- Zhang, Y.Z. Novel 2019 Coronavirus Genome. Virological. Available online: http://virological.org/t/novel-2019-coronavirus-genome/319 (accessed on 14 January 2021).
- World Health Organization. Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases. Available online: https://apps.who.int/iris/handle/10665/330676 (accessed on 14 January 2021).
- General Office of the National Health and Health Commission Office of the NA of TCM. Diagnosis and Treatment of Pneumonitis with a New Type of Coronavirus Infection (Trial Version 7). Available online: https://www.who.int/docs/default-source/wpro---documents/countries/china/covid-19-briefing-nhc/1-clinical-protocols-for-the-diagnosis-and-treatment-of-covid-19-v7.pdf?sfvrsn=c6cbfba4_2 (accessed on 3 January 2021).
- National Medical Products Administration. New Novel Coronavirus Detection Products Approved by the National Medical Products Administration. Available online: http://www.nmpa.gov.cn/WS04/CL2056/375802.html (accessed on 15 May 2020).
- United States Food and Drug Administration. Available online: https://www.fda.gov/medical-devices/emergency-use-authorizations-medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices (accessed on 15 May 2020).
- Arevalo-Rodriguez, I.; Buitrago-Garcia, D.; Simancas-Racines, D.; Zambrano-Achig, P.; Del Campo, R.; Ciapponi, A.; Sued, O.; Martinez-García, L.; Rutjes, A.W.; Low, N.; et al. False-negative results of initial RT-QPCR assays for COVID-19: A systematic review. PLoS ONE 2020, 15, 0242958. [Google Scholar] [CrossRef]
- Lippi, G.; Simundic, A.M.; Plebani, M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin. Chem. Lab. Med. 2020, 50, 1070–1076. [Google Scholar] [CrossRef]
- Li, D.; Wang, D.; Dong, J.; Wang, N.; Huang, H.; Xu, H.; Xia, C. False-Negative Results of Real-Time Reverse-Transcriptase Polymerase Chain Reaction for Severe Acute Respiratory Syndrome Coronavirus 2: Role of Deep-Learning-Based CT Diagnosis and Insights from Two Cases. Korean J. Radiol. 2020, 21, 505–508. [Google Scholar] [CrossRef]
- McKinsey and Company. COVID-19: Overcoming Supply Shortages for Diagnostic Testing. Available online: http://www.mckinsey.com/industries/pharmaceuticals-and-medical-products/our-insights/covid-19-overcoming-supply-shortages-for-diagnostic-testing?cid=eml-web (accessed on 8 July 2020).
- Palka-Santini, M.; Cleven, B.E.; Eichinger, L.; Krönke, M.; Krut, O. Large scale multiplex PCR improves pathogen detection by DNA microarrays. BMC Microbiol. 2009, 9, 1–16. [Google Scholar] [CrossRef]
- Mou, X.; Ali, Z.; Li, B.; Li, T.; Yi, H.; Dong, H.; He, N.; Deng, Y.; Zeng, X. Multiple genotyping based on multiplex PCR and microarray. Chin. Chem. Lett. 2016, 27, 1661–1665. [Google Scholar] [CrossRef]
- Zheng, Z.; Wu, Y.; Yu, X.; Shang, S. DNA Microarray Technology for Simultaneous Detection and Species Identification of Seven Human Herpes Viruses. J. Med. Virol. 2008, 80, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Damin, F.; Galbiati, S.; Soriani, N.; Burgio, V.; Ronzoni, M.; Ferrari, M.; Chiari, M. Analysis of KRAS, NRAS and BRAF mutational profile by combination of in-tube hybridization and universal tag-microarray in tumor tissue and plasma of colorectal cancer patients. PLoS ONE 2018, 13, e0207876. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, S.; Damin, F.; Ferraro, L.; Soriani, N.; Burgio, V.; Ronzoni, M.; Gianni, L.; Ferrari, M.; Chiari, M. Microarray approach combined with ddPCR: An useful pipeline for the detection and quantification of circulating tumor DNA mutations. Cells 2019, 8, 769. [Google Scholar] [CrossRef]
- Galbiati, S.; Monguzzi, A.; Damin, F.; Soriani, N.; Passiu, M.; Castellani, C.; Natacci, F.; Curcio, C.; Seia, M.; Lalatta, F.; et al. COLD-PCR and microarray: Two independent highly sensitive approaches allowing the identification of fetal paternally inherited mutations in maternal plasma. J. Med. Genet. 2016, 53, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Ozkumur, E.; Yalcin, A.; Cretich, M.; Lopez, C.A.; Bergstein, D.A.; Goldberg, B.B. Quantification of DNA and protein adsorption by optical phase shift. Biosens. Bioelectron. 2009, 25, 167–172. [Google Scholar] [CrossRef]
- Cretich, M.; di Carlo, G.; Longhi, R.; Gotti, C.; Spinella, N.; Coffa, S.; Galati, C.; Renna, L.; Chiari, M. High sensitivity protein assays on microarray silicon slides. Anal. Chem. 2009, 81, 5197–5203. [Google Scholar] [CrossRef] [PubMed]
- Pirri, G.; Damin, F.; Chiari, M.; Bontempi, E.; Depero, L.E. Characterization of a polymeric adsorbed coating for DNA microarray glass slides. Anal. Chem. 2004, 76, 1352–1358. [Google Scholar] [CrossRef]
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X.; et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes. Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef]
- Falzone, L.; Musso, N.; Gattuso, G.; Bongiorno, D.; Palermo, C.I.; Scalia, G.; Libra, M.; Stefani, S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020, 46, 957–964. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-QPCR Diagnostic Panel. Available online: https://www.fda.gov/media/134922/download (accessed on 18 January 2021).
- Mak, G.C.K.; Cheng, P.K.C.; Lau, S.S.Y.; Wong, K.K.Y.; Lau, C.S.; Lam, E.T.K.; Chan, R.C.W.; Tsang, D.N.C. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 129. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Sakai-Tagawa, Y.; Koga, M.; Akasaka, O.; Nakachi, I.; Koh, H.; Maeda, K.; Adachi, E.; Saito, M.; Nagai, H.; et al. Comparison of Rapid Antigen Tests for COVID-19. Viruses 2020, 12, 1420. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. COVID-19 Diagnostic Test Accuracy Group. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020. [Google Scholar] [CrossRef]
- Konrad, R.; Eberle, U.; Dangel, A.; Treis, B.; Berger, A.; Bengs, K.; Fingerle, V.; Liebl, B.; Ackermann, N.; Sing, A. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Eurosurveillance 2020, 25, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Han, C.M.; Catoe, D.; Munro, S.A.; Khnouf, R.; Snyder, M.P.; Santiago, J.G.; Salit, M.L.; Cenik, C. Simultaneous RNA purification and size selection using on-chip isotachophoresis with an ionic spacer. Lab Chip 2019, 19, 2741–2749. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; Han, J. One-Step Nucleic Acid Purification and Noise-Resistant Polymerase Chain Reaction by Electrokinetic Concentration for Ultralow-Abundance Nucleic Acid Detection. Angew. Chem. Int. Edit. 2020, 59, 10981–10988. [Google Scholar] [CrossRef] [PubMed]
- Damin, F.; Galbiati, S.; Ferrari, M.; Chiari, M. DNA microarray-based solid-phase PCR on copoly (DMA–NAS–MAPS) silicon coated slides: An example of relevant clinical application. Biosens. Bioelectron. 2016, 78, 367–373. [Google Scholar] [CrossRef]
- Wang, X.; Yao, H.; Xu, X.; Zhang, P.; Zhang, M.; Shao, J.; Xiao, Y.; Wang, H. Limits of detection of six approved RT-QPCR kits for the novel SARS-coronavirus-2 (SARS CoV-2). Clin. Chem. 2020, 66, 977–979. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Ardebili, A. Real-time RT-QPCR in COVID 19 detection: Issues affecting the results. Expert. Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef]





| Nasopharyngeal Swabs | N1 qPCR (Ct) 1 | N2 qPCR (Ct) 1 |
|---|---|---|
| S1 | 20.61 | 20.45 |
| S2 | 21.83 | 29.31 |
| S3 | N/A 2 | N/A 2 |
| S4 | 38.49 | N/A 2 |
| S5 | 35.86 | N/A 2 |
| Sample Id | N1 qPCR (Ct) 1 | N2 qPCR (Ct) 1 | CovidArray |
|---|---|---|---|
| N017 | 34.66 | 39.35 | POSITIVE |
| N053 | N/Ab | N/Ab | NEGATIVE |
| B001 | 34.74 | 35.16 | POSITIVE |
| N141 | 14.48 | 12.83 | POSITIVE |
| N051 | N/A 2 | N/A 2 | NEGATIVE |
| B210 | 33.24 | 42.5 | POSITIVE |
| B243 | 25.82 | 39.29 | POSITIVE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damin, F.; Galbiati, S.; Gagliardi, S.; Cereda, C.; Dragoni, F.; Fenizia, C.; Savasi, V.; Sola, L.; Chiari, M. CovidArray: A Microarray-Based Assay with High Sensitivity for the Detection of Sars-Cov-2 in Nasopharyngeal Swabs. Sensors 2021, 21, 2490. https://doi.org/10.3390/s21072490
Damin F, Galbiati S, Gagliardi S, Cereda C, Dragoni F, Fenizia C, Savasi V, Sola L, Chiari M. CovidArray: A Microarray-Based Assay with High Sensitivity for the Detection of Sars-Cov-2 in Nasopharyngeal Swabs. Sensors. 2021; 21(7):2490. https://doi.org/10.3390/s21072490
Chicago/Turabian StyleDamin, Francesco, Silvia Galbiati, Stella Gagliardi, Cristina Cereda, Francesca Dragoni, Claudio Fenizia, Valeria Savasi, Laura Sola, and Marcella Chiari. 2021. "CovidArray: A Microarray-Based Assay with High Sensitivity for the Detection of Sars-Cov-2 in Nasopharyngeal Swabs" Sensors 21, no. 7: 2490. https://doi.org/10.3390/s21072490
APA StyleDamin, F., Galbiati, S., Gagliardi, S., Cereda, C., Dragoni, F., Fenizia, C., Savasi, V., Sola, L., & Chiari, M. (2021). CovidArray: A Microarray-Based Assay with High Sensitivity for the Detection of Sars-Cov-2 in Nasopharyngeal Swabs. Sensors, 21(7), 2490. https://doi.org/10.3390/s21072490