1. Introduction
The handling of a crime scene is an important part of successful and dynamic criminal investigations. Forensic science deals with true crime casework for the collection, detection, and analysis of evidence material. Different traces that are found in crime casework can be very important. A schematic search, analysis, and conclusions from these traces make them valuable in court investigations [
1,
2,
3,
4]. One of the most important forms of forensic shreds of evidence found at a crime scene is body fluids. Blood is a valuable and common body fluid found at violent crime scenes [
5]. The analysis of bloodstain patterns and the age determination of bloodstains are interesting areas of forensic casework that can lead to identifying suspects [
6,
7].
In the case of stain detection at a crime scene, the first challenge is to develop a technique for the confirmation of a stain as a bloodstain. This is because a bloodstain can be comparable to other substances in terms of color and appearance on different substrates on visual inspection [
8]. Though Deoxyribonucleic Acid (DNA) can be used to identify a suspect, however, DNA analysis is time-consuming and expensive. The presence of false positives like a stain of brown paint may particularly lead to a waste of resources and time. Therefore, true blood stains should be selected for subsequent DNA analysis.
Several presumptive tests are used for the identification of stains as bloodstains against other confusing substances. These tests include chemical methods, for instance, Kastle–Meyer (KM), Leucomalachite Green (LMG), Benzidine [
9], and Luminol [
10]. The KM test is also known as the phenolphthalein test and it may produce false positives [
11]. Although it is not sensitive, it can detect blood after a dilution ratio of 1 in 10,000. LMG is also sensitive as KM with a 1 in 10,000 dilution ratio [
12]. The luminol test is very sensitive as compared to the KM test and produces fewer false positives. Moreover, luminol test requires a dark environment [
13,
14].
As all these tests are presumptive, therefore, confirmatory tests are required to reduce false positives. These confirmatory tests include spectroscopic, chromatographic, microscopic, and crystal tests. All these aforementioned methods may use chemicals or require sample preparation that can be problematic for subsequent analysis like pattern and DNA analysis. Moreover, a small amount of biological traces are available in some cases. Therefore, these tests can destroy the original context [
15,
16].
Therefore, the forensic work is captivated by non-destructive methods for the identification of shreds of evidence for the recent period [
17]. Different non-contact techniques have been used in the forensic area. Practitioners are using different spectroscopic techniques like Raman, Reflectance, Electron Paramagnetic Resonance (EPR), Nuclear Magnetic Resonance (NMR), and Infrared (IR) spectroscopies like Attenuated Total Reflectance Fourier Transform IR spectroscopy (ATR-FTIR). IR and Raman-based techniques provide promising results for bloodstain identification [
18].
For instance, Bremmer et al. used reflectance spectroscopy in the visible region to identify blood based on the correlation coefficient [
19]. Edelman et al. used spectroscopy in NIR and found difficulty in blood identification in the visible region for colored substrates due to high absorbance. The authors did not include any non-blood substance containing protein to avoid false positives. Moreover, the method was best suitable for dried samples due to the water peak in the NIR [
20]. Pereira et al. identified human blood through the evaluation of different supervised pattern recognition techniques with NIR spectroscopy [
21]. Morillas et al. used a portable NIR spectrometer for bloodstain identification. The authors observed good results with 81–94% accuracy with different classification models [
22].
Likewise, Raman spectroscopy was used to distinguish human and non-human species, for gender identification and to discriminate between infant and adult blood donors. Moreover, ATR-FTIR spectroscopy was used in different forensic applications [
18,
23,
24], but IR technologies can be used only for IR active substances. Though Raman technology has more promising results than IR, in forensic applications, however, its accuracy of forensic work degrades for dried-up samples due to the degradation of blood structures with powered laser lights. Moreover, complications are found in the case of overlaying weak bands while dealing with Raman signals. This may require expertise and significant analysis [
25]. All these spectroscopy techniques are limited to provide the spectral information of the whole specimen instead of spatial information of the whole area under observation. Spatial information helps to extract information of samples with different shapes on different backgrounds. For this purpose, spectral imaging has been used widely, which combines the benefits of both imaging and spectroscopy.
Irrespective to Raman spectroscopy, Hyperspectral Imaging (HSI) with different substrates especially dark and patterned has been proposed in [
26]. The authors of [
26] found that the bloodstains were visible only at few wavelengths in the visible and NIR range. Edelman et al. demonstrated the visualization of bloodstains on black backgrounds using chemo-metrical techniques with visible HSI. The authors also concluded that the bloodstain identification task with other substances was tedious for dark substrates in this range [
27]. B. Li et al. proposed a novel approach for bloodstain identification using Soret peak at 415 nm in visible HSI [
28]. Cadd et al. used the same criteria of correlation coefficient with the soret band for the identification of blooded fingerprints on white tiles [
29] and different colored tiles [
30] in their future studies. Cadd et al. extended the work with a chemical enhancement of Acid Black 1 for the first time to detect blood-stained fingerprint [
31]. In all of the above, temperature and humidity must be maintained to avoid blue spectral shift [
32]. Moreover, the authors reported high accuracy with a soret band instead of weak
and
absorption bands.
Irrespective of the works discussed above, in this work, HSI technology has been used for non-contact identification of blood traces, thus limiting the problem of destruction and contamination of these traces. HSI gives both spatial and spectral information of material under observation [
33]. Hence, fast data acquisition, less expected human error, and no sample preparation lessens the load of work in labs and helps in further analysis after quick identification. In the modern era, HSI finds its applications in food quality assessment, medical imaging, security and defense, remote sensing [
34,
35] field, and artwork authentication [
36].
In a nutshell, in this work, a bloodstain is identified against eight different blood resembling substances using the HSI system in the 397–1000 nm range. The blood samples from three donors were imaged, using three different substrates (white cotton fabric, white tile, and PVC wall sheet) with eight different non-blood items (ketchup, rust acrylic paint, red acrylic paint, brown acrylic paint, red nail polish, rust nail polish, fake blood, and red ink). Savitzky Golay derivative has been used to enhance the features of the blood spectrum. The important wavelengths (bands) were used to train and test five different classification models. Finally, a blind experiment has been performed with a blood sample of the fourth blood donor and different blood resembling substances on each substrate for further validation of the proposed methodology.
2. Materials and Methods
This section summarizes the sample preparation, hardware system, data acquisition, data pre-processing, and identification criteria with different classification models.
2.1. HSI System
HSI system used in this study includes an FX-10 (Specim, Spectral Imaging Ltd, Finland) Hyperspectral camera, equipped with a lens from Scheiner and a line scanner. The setup contained three halogen lamps, a moving platform for scanning, and a camera mounting plate with an adjustable height. A serial communication port was used to connect the scanner directly to a laptop where data was dumped by a software Lumo Scanner. GigE-Vision was interfaced with a camera and laptop to transfer captured data. Three types of raw files including dark reference, white reference, and sample were obtained. For dark and white references, 100 frames were acquired by closing the shutter and using a white tile, respectively. The HSI system is capable of capturing a hypercube of a size of 1000 × 512 × 224 in the visible NIR range between 397–1003 nm with an average subsampling of 2.7 nm.
2.2. Sample Preparation
In this study, human blood is used for identification purposes against different blood resembling substances including ketchup, rust acrylic paint, red acrylic paint, brown acrylic paint, red nail polish, rust nail polish, fake blood, and red ink. The blood was stained on different substrates directly from the fingertips after the informed consent of volunteers during the whole experimentation. For this purpose, the ACCU-CHECK® safe lancing device was used with sterilized needles. The needle was disposed of after each blood sample deposition. Before blood sample deposition, all substrates were cleaned with distilled water. The stain size of each blood and non-blood sample were tried to be kept within a diameter of about 1 cm but the number of pixels within one stain could be varied. To apply the same number of stains for each surface, a different number of pieces of each substrate were used according to their available surface area.
The HSI cubes (blood and non-blood samples) were captured for up to three days in order to include dehydrated (aging) samples. The samples were placed to dry for about one hour before imaging on the first day. Then the samples were kept at room temperature in an airtight box. The observed averaged temperature and humidity were 39 °C and 40%, respectively. The precise information about the number of samples (HSI cubes) is shown in
Table 1. The details related to number of pixel spectra of all stains and their distribution in different sets are shown in
Table 2 and
Table 3.
The sample deposition information for three substrates (white cotton fabric, white tile, and wall sheet) is given below:
Three bloodstains were deposited by each of the three donors, creating a total of nine stains on each substrate. These samples were imaged for three days so making a dataset of 27 samples (HSI cubes) for each substrate. These blood samples together with non-blood samples were used for model training and testing;
For blood resembling substances, two stains of each substance (ketchup, rust acrylic paint, red acrylic paint, brown acrylic paint, red nail polish, rust nail polish, fake blood, and red ink) were deposited on each substrate and imaged for up to three days as similar to the blood samples. A total of 48 samples (HSI cubes) of 8 different blood resembling substances along with 27 bloodstains were used for training and testing of the models for each substrate;
Blind Trial: For the final evaluation and validation of trained/tested models, a blind experiment was also conducted on entirely unseen blood samples (HSI cubes). These samples were collected as; each substrate was stained with two blood samples of different aging, from another donor along with four non-blood samples (6 HSI cubes). These HSI cubes are of size 410 × 512 × 224 as compared to the previous experiments which were conducted on 100 × 100 × 224.
2.3. Spectral Reflectance
The samples (blood, non-blood stains) were placed on the moving platform of the HSI system. A white tile of 99.9% reflectance placed on a moving platform was used as a white reference. The speed of the platform was set to 11.72 mm/s. The frame rate was 50 Hz and the exposure time was 16 ms. The camera was adjusted at a height of 15 cm. These settings were kept constant for the entire data collection. As the size of each substrate was different, therefore, the number of frames of hypercube was set according to the size of different substrates.
The hyperspectral camera records the radiance of the specimen. The recorded radiance suffers from different factors like a spectrum of the illumination source, incident angle with a specimen, atmospheric effects, shadowing, and sensor effects. Therefore, it is necessary to convert radiance into spectral reflectance with the removal of different factors. For this purpose, two calibration targets, white reference and dark reference, with a wide brightness difference, were used to calculate reflectance from encoded sample radiance by using the Empirical Line Method [
37]. Path radiance and shadowing effects are also eliminated with Empirical Line Method. The linear equation is used to calculate spectral reflectance from encoded radiance for each spectral band of a hypercube.
where
is encoded radiance of sample while
and
are captured dark and white reference frames.
2.4. Pre-Processing
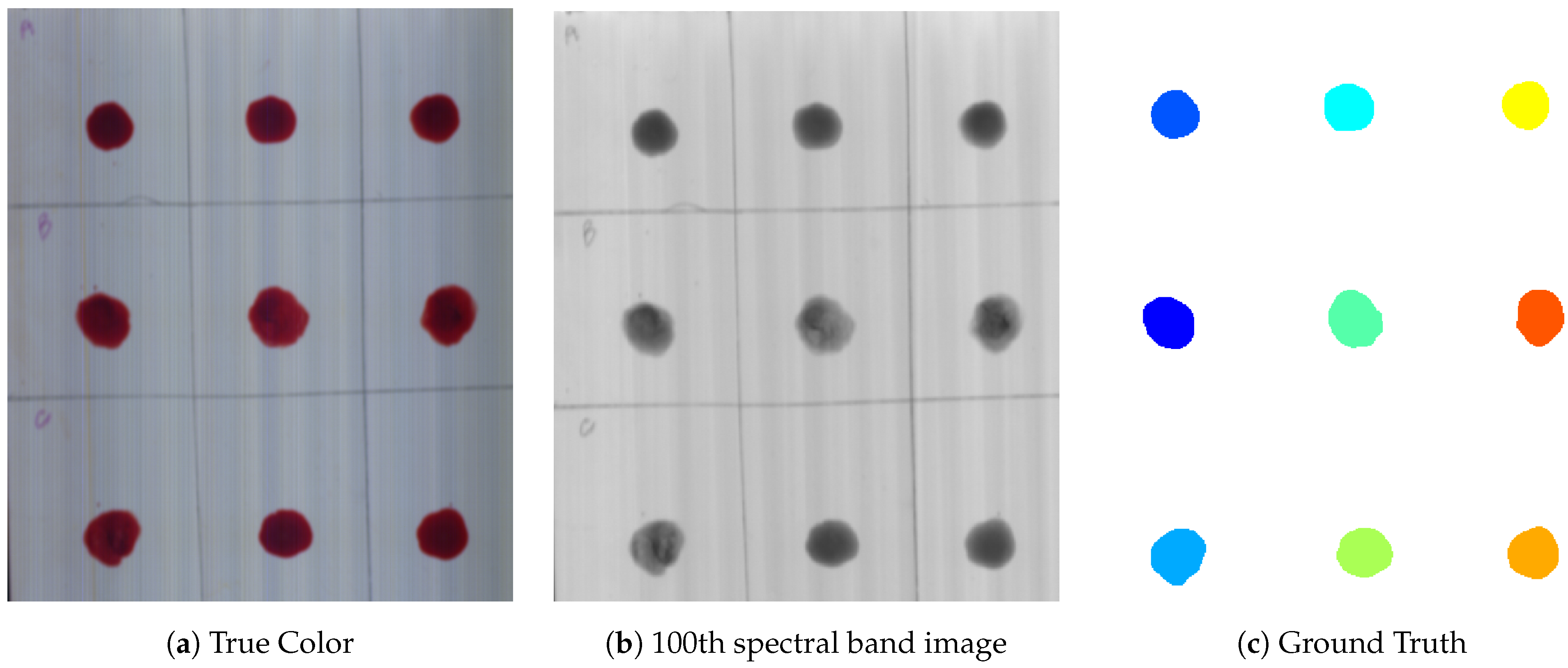
The pre-processing was divided into two parts, i.e., spatial and spectral pre-processing. Image pre-processing techniques have been applied in order to extract a Region of Interest (ROI) from an image. For ROI, a Hyperspectral reflectance image of a data cube of 100 × 100 pixels for each blood and non-blood sample was cropped. Then, thresholding was done at 540 nm. After extracting the mask, it was multiplied with the whole hypercube to extract the spectral information of pixels related to ROI as shown in
Figure 1. To remove the noise, Standard Normal Variate (SNV), Multiplicative Scatter Correction (MSC), Smoothing with Savitzky Golay method, and averaging filter were used [
21,
38,
39,
40].
Based on visual inspection, the Savitzky Golay smoothing filter performed well for noise removal. To deal with pixel information in an HSI cube, a derivative is a helpful technique to detect pixel variations, subtle characteristics, and weak absorption bands. First-order derivatives can be used to remove spectral shifts. However, in this work, higher-order derivatives are used to extract overlapping spectral features and for background elimination [
41]. Higher-order derivatives cause a reduction in signal-to-noise ratio (SNR). Savitzky Golay filter is one of the mathematical derivative methods that are better than others due to its soothing property [
42,
43,
44,
45]. The Savitzky Golay filter is based on polynomial fitting on data points depending upon window size. The solution of a polynomial is found by the least square minimization. The polynomial order is fixed depending on the derivative order to be calculated [
46]. In this study, Savitzky Golay’s second order derivative with a 13-point window and third-order polynomial was used to remove spectral shifts due to aging, irrelevant noise, different donor samples, and pixel variations. The derivative of aged samples also highlighted the subtle dips to make the blood spectrum homogeneous.
2.5. Identification Criteria of Blood from other Red Substances
Hemoglobin is an important component of blood. Dried-up bloodstains contain about 97% of hemoglobin components. Oxyhemoglobin, meta-hemoglobin, and hemi-chrome are important hemoglobin derivatives in vitro reactions [
47]. The spectral properties of hemoglobin derivatives are used in this study to develop identification criteria. The mean reflectance spectra and their derivatives of blood samples are observed against different red-colored substances that could be confused with blood. In the visible region, two peaks
and
are found due to oxyhemoglobin at wavelengths 577 nm and 540 nm respectively as shown in
Figure 2. With aging, the dips become less prominent on visual inspection in a spectral signature.
Moreover, pixel-wise reflectance suffered from spectral noises may cause spectral shifts. The steepness in the curve of blood is observed from 600–650 nm, which is due to the formation of hemi-chrome and meta-hemoglobin, moreover, with aging, this steepness also decreases [
48]. As mentioned earlier, the Savitzky Golay derivative is used to make dips and peaks of aged blood samples and non-blood samples more prominent. Moreover, it also highlighted a prominent change in the blood derivative spectrum against a non-blood derivative spectrum between 470–770 nm. In the derivative spectrum, redundant information is ignored, therefore, appropriate spectral bands are selected based on derivative spectra for classification models.
5. Experimental Settings and Results
For experimental evaluation, several statistical tests have been conducted including but not limited to Kappa
, overall accuracy, sensitivity, specificity, F1-score, precision, and recall rate. All these evaluation metrics are calculated using the following mathematical formulations.
where
where
TP and
FP are true and false positive, and
TN and
FN are true and false negative computed from the confusion matrix.
Experiments have been conducted using five different types of classifiers, i.e., Support Vector Machine (SVM) [
49], K Nearest Neighbors (KNN) [
49], Artificial Neural Network (ANN) [
50], Decision Trees (DT) [
51], and Random Forest (RF) [
51]. The tuning parameters for all the aforementioned classifiers are as follows: In the case of ANNs, a dense network with three hidden layers, each containing 30 neurons, was used. The number of layers and nodes were selected by the hit and trial method. In order to increase sensitivity and specificity, the number of epochs was increased up to 50. SVM is trained using Linear and RBF kernel functions and rest of the parameters are left default. KNN was tuned between [1, 20] nearest neighbors iteratively. DT was trained using entropy criterion with 10 times depth and finally, and RF was trained using 500 estimators.
The tuning parameters of all the above-said classifiers were explored very carefully in the first few experiments (as listed above) and those that provided the best accuracy were chosen. To avoid bias, all the listed experiments are carried out in the same settings on the same machine. Before the experiments, we performed the necessary normalization between [0, 1], and all the experiments are carried out using Matlab 2021a installed on an Intel inside Core i3-4030U CPU 1.90 GHz with 12 GB RAM.
As discussed in
Section 4, classification was performed with two separate data sets. For feature reduction, Principle Component Analysis (PCA) was performed on each sample data with a selection of all wavelengths which significantly reduces the performance of SVM even with different kernels functions. Then, the results were observed with a second-order derivative spectrum with two spectral ranges i.e., 397–770 nm and 470–770 nm. Similar results have been drawn with both spectral ranges, therefore, a 470–770 nm spectral range was selected to train models with a fewer number of features which improves the generalization performance. It has been observed that all the classifiers performed with 100% accuracy and statistical significance for all blood samples as given in
Table 4 and
Table 5.
Similarly, blood vs. blood (donor A, B, and C as a separate class) vs. 8 other non-blood samples (protein-based ketchup, rust acrylic paint, red acrylic paint, brown acrylic paint, red nail polish, rust nail polish, fake blood, and red ink) on three different substrates (white cotton fabric, white tile, and PVC wall sheet) as multi-class classification results have also been carried to show the performance of our proposed pipeline. Moreover, the ageing of all these results have also been discussed in
Table 6,
Table 7 and
Table 8, respectively.
7. Comparison with State-of-the-Art Methodologies
In the literature, PCA is a commonly used method for feature reduction. PCA is used to map high-dimensional data to lower dimensions while taking a linear combination of original data with orthogonal vectors known as Principal Components (PCs) [
52]. The top PCs of transformed data carry as much variation as possible. To strengthen our proposed pipeline, feature reduction has been performed with PCA instead of derivatives. We have trained our models with the first three PCs for comparison.
Moreover, study [
21] evaluated different supervised pattern recognition methods for blood identification. This study followed Standard Normal Variate (SNV) and Normalization in the pre-processing step. We have followed their two pipelines with Soft Independent Modeling of Class Analogy (SIMCA) and Partial-Least Squares Discriminant Analysis (PLS-DA) models to compare with our results. The number of PCs selected for both models is 3 and 2, respectively.
Table 9 presents the results of these methodologies against our proposed pre-processing method and derivative-based feature selection. These results have been drawn with the same data set used in this study.
The results have been presented for the identification of blood against eight different blood resembling substances using HSI technology in the visible region. These blood and non-blood samples were deposited on different substrates that are commonly discussed in forensic applications. These substrate materials included white cotton fabric, white tile, and a PVC wall sheet.
In order to reconstruct old crime scenes, a criterion of aging for three days was set. Therefore, the images were captured for up to three days. The reflectance spectra of all blood and non-blood samples were analyzed visually and the identification criteria are set based on blood heme-components. First, PCA was used for feature reduction from the reflectance spectrum and classification results were observed. To observe reduced performance with PCA, the distribution of transformed data with aged samples is also analyzed. The distribution is randomly scattered due to the change in the chemical composition of different substances. Then Savitzky Golay derivative filter is used in the pre-treatment step to enhance features of aged samples. Therefore, derivative-based important spectral lines are selected while discarding redundant information. The performance of different models used in this study increased to 100% with derivative-based features. The proposed methodology provided the considerable potential to identify blood with Hyperspectral imaging.