Multi-Task Model for Esophageal Lesion Analysis Using Endoscopic Images: Classification with Image Retrieval and Segmentation with Attention
Abstract
:1. Introduction
- (1)
- We proposed a novel multi-task deep learning model for automatic esophageal lesion analysis. It can synchronously achieve multiple tasks, including classification and segmentation for esophageal lesions.
- (2)
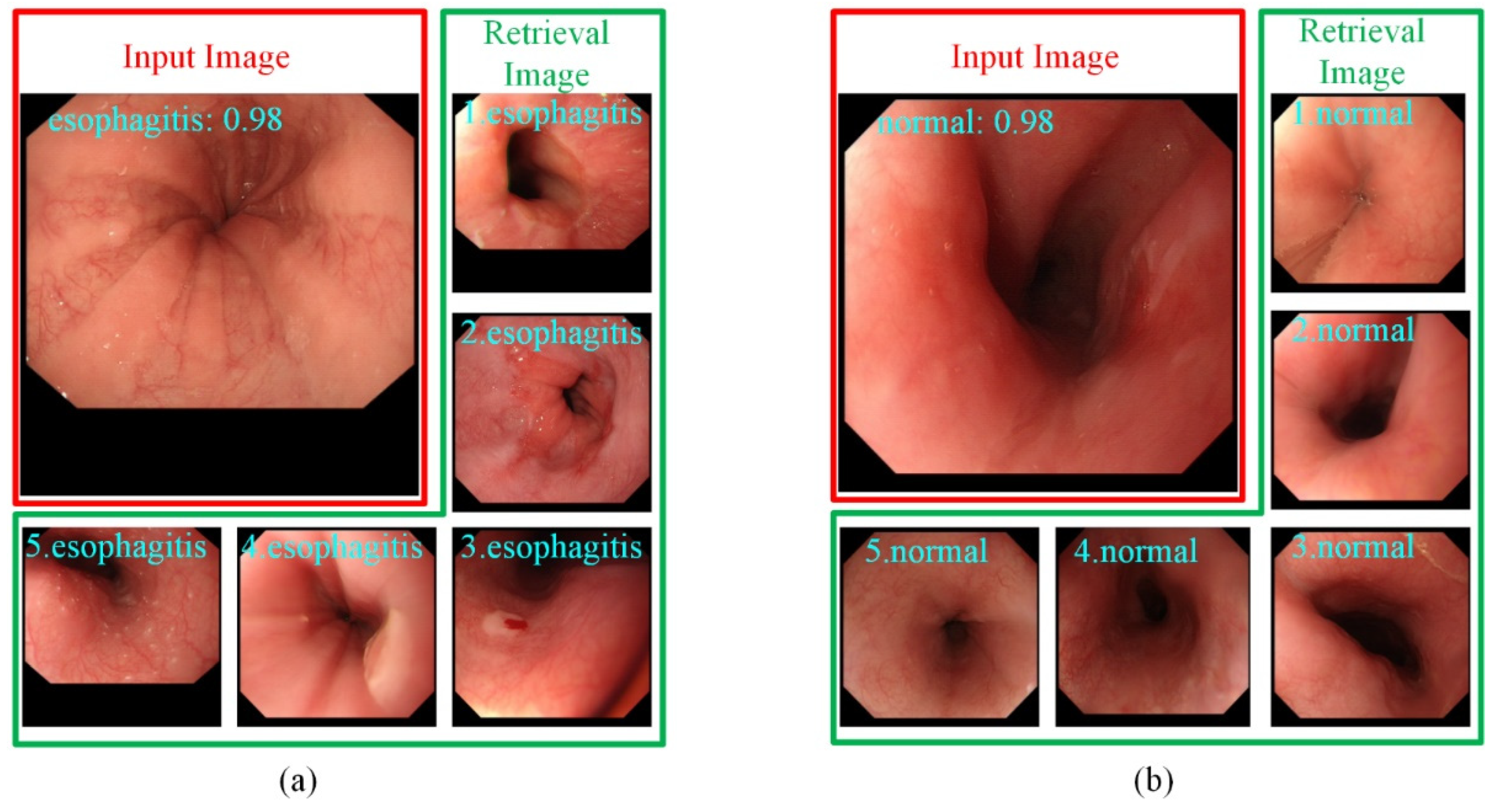
- To provide endoscopists with more supporting information in classification, we built a retrieval module on the classification branch to assign a confidence for each prediction result. Classification and retrieval can be optimized at the same time without affecting each other.
- (3)
- To improve the performance of esophageal cancer segmentation, we designed a mutual attention module in the segmentation task that can generate weight matrices from different features and guide each other to obtain diversified features.
- (4)
- The experiments show that the proposed model is better than other similar methods and can effectively help endoscopists improve the accuracy of a diagnosis.
2. Related Works
2.1. Esophagus Classification
2.2. Esophagus Segmentation
2.3. Multi-Task in Medical Image Analysis
3. Proposed Methods
3.1. Network Architecture of The Proposed Multi-Task Deep Learning Model
3.2. Classification and Segmentation Tasks
3.2.1. The Classification Task
3.2.2. The Segmentation Task
4. Experiments and Discussion
4.1. Dataset
4.2. Evaluation Metric
4.3. The Classification Results
4.4. The Segmentation Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Rice, T.W.; Ishwaran, H.; Hofstetter, W.; Kelsen, D.; Apperson-Hansen, C.; Blackstone, E.H. Recommendations for pathologic staging (pTNM) of cancer of the esophagus and esophagogastric junction for the 8th edition AJCC/UICC staging manuals. Dis. Esophagus 2016, 29, 897–905. [Google Scholar] [CrossRef] [Green Version]
- Ezoe, Y.; Muto, M.; Uedo, N.; Doyama, H.; Yao, K.; Oda, I.; Kaneko, K. Magnifying narrowband imaging is more accurate than conventional white-light imaging in diagnosis of gastric mucosal cancer. Gastroenterology 2011, 141, 2017–2025. [Google Scholar] [CrossRef] [Green Version]
- Barbeiro, S.; Libanio, D.; Castro, R.; Dinis-Ribeiro, M.; Pimentel-Nunes, P. Narrow-band imaging: Clinical application in gastrointestinal endoscopy. GE Port. J. Gastroenterol. 2018, 26, 40–53. [Google Scholar] [CrossRef]
- Pennazio, M. Capsule endoscopy: Where are we after 6 years of clinical use? Dig. Liver Dis. 2006, 38, 867–878. [Google Scholar] [CrossRef]
- Mannath, J.; Ragunath, K. Role of endoscopy in early oesophageal cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 720–730. [Google Scholar] [CrossRef]
- Du, W.; Rao, N.; Liu, D.; Jiang, H.; Luo, C.; Li, Z.; Gan, T.; Zeng, B. Review on the applications of deep learning in the analysis of gastrointestinal endoscopy images. IEEE Access 2019, 7, 142053–142069. [Google Scholar] [CrossRef]
- Ameri, A. A deep learning approach to skin cancer detection in dermoscopy images. J. Biomed. Phys. Eng. 2020, 10, 801–806. [Google Scholar] [CrossRef]
- Liskowski, P.; Krawiec, K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans. Med. Imaging 2016, 35, 2369–2380. [Google Scholar] [CrossRef]
- Arif, M.; Schoots, I.; Tovar, J.; Bangma, C.; Krestin, G.; Roobol, M.; Nieesen, W.; Veenland, J. Clinically significant prostate cancer detection and segmentation in low-risk patients using a convolutional neural network on multi-parametric MRI. Eur. Radiol. 2020, 30, 6582–6592. [Google Scholar] [CrossRef]
- Luo, H.; Xu, G.; Li, C.; He, L.; Luo, L.; Wang, Z.; Jing, B.; Deng, Y.; Jin, Y.; Li, B.; et al. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: A multicentre, case-control, diagnostic study. Lancet 2019, 20, 1645–1654. [Google Scholar] [CrossRef]
- Nakagawa, K.; Ishihara, R.; Aoyama, K.; Ohmori, M.; Nakahira, H.; Matsuura, N.; Shichijio, S.; Nishida, T.; Yamada, T.; Yamaguchi, S.; et al. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest. Endosc. 2019, 90, 407–414. [Google Scholar] [CrossRef]
- Cao, R.; Pei, R.; Ge, N.; Zheng, C. Clinical target volume auto-segmentation of esophageal cancer for radiotherapy after radical surgery based on deep learning. Technol. Cancer Res. Treat. 2021, 20, 1–11. [Google Scholar] [CrossRef]
- Guo, L.; Xiao, X.; Wu, C.; Zeng, X.; Zhang, Y.; Du, J.; Bai, S.; Zhang, Z.; Li, Y.; Wang, X. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest. Endosc. 2020, 91, 41–51. [Google Scholar] [CrossRef]
- Caruana, R. Multitask learning. Mach. Learn. 1998, 27, 95–133. [Google Scholar]
- Misra, I.; Shrivastava, A.; Gupta, A.; Hebert, M. Cross-Stitch Networks for Multi-Task Learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Kokkinos, I. Ubernet: Training a Universal Convolutional Neural Network for Low-, Mid-, and High-Level Vision Using Diverse Datasets and Limited Memory. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Caruana, R. Multitask Learning: A Knowledge Based Source of Inductive Bias. In Proceedings of the Tenth International Conference on Machine Learning, Amherst, MA, USA, 27–29 July 1993. [Google Scholar]
- Münzenmayer, C.; Kage, A.; Wittenberg, T.; Mühldorfer, S. Computer-assisted diagnosis for precancerous lesions in the esophgus. Methods Inf. Med. 2009, 48, 324–330. [Google Scholar]
- Riaz, F.; Silva, F.; Ribeiro, M.; Coimbra, M. Invariant gabor texture descriptors for classification of gastroenterology images. IEEE Trans. Biomed. Eng. 2012, 59, 2893–2904. [Google Scholar] [CrossRef]
- Yeh, J.Y.; Wu, T.; Tsai, W.J. Bleeding and ulcer detection using wireless capsule endoscopy images. J. Softw. Eng. Appl. 2014, 7, 422–432. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Gan, T.; Rao, N.; Xing, Y.; Zheng, J.; Li, S.; Luo, C.; Zhou, Z.; Wan, Y. Identification of lesion images from gastrointestinal endoscope based on feature extraction of combinational methods with and without learning process. Med. Image Anal. 2016, 32, 281–294. [Google Scholar] [CrossRef]
- Kumagai, Y.; Takubo, K.; Kawada, K.; Aoyama, K.; Endo, Y.; Ozawa, T.; Hirasawa, T.; Yoshio, T.; Ishihara, S.; Fujishiro, M.; et al. Diagnosis using deep-learning artificial intelligence based on the endocytoscopic observation of the esophagus. Esophagus 2019, 16, 180–187. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C.; Bai, J.; Liao, G. Fine-tuning pre-trained convolutional neural networks for gastric precancerous disease classification on magnification narrow-band imaging images. Neurocomputing 2020, 392, 253–267. [Google Scholar] [CrossRef]
- Du, W.; Rao, N.; Dong, C.; Wang, Y.; Hu, D.; Zhu, L.; Zeng, B.; Gan, T. Automatic classification of esophageal disease in gastroscopic images using an efficient channel attention deep dense convolutional neural network. Biomed. Opt. Express 2021, 12, 3066–3081. [Google Scholar] [CrossRef]
- Igarashi, S.; Sasaki, Y.; Mikami, T.; Sakuraba, H.; Fukuda, S. Anatomical classification of upper gastrointestinal organs under various image capture conditions using AlexNet. Comput. Biol. Med. 2020, 124, 103950. [Google Scholar] [CrossRef]
- Fieselmann, A.; Lautenschläger, S.; Deinzer, F.; Matthias, J.; Poppe, B. Esophagus segmentation by spatially-constrained shape interpolation. In Proceedings of the Bildverarbeitung für die Medizin 2008: Algorithmen–Systeme–Anwendungen, Proceedings des Workshops, Berlin, Germany, 6–8 April 2008; pp. 247–251. [Google Scholar]
- Feulner, J.; Zhou, S.; Cavallaro, A.; Seifert, S.; Hornegger, J.; Comaniciu, D. Fast automatic segmentation of the esophagus from 3D CT data using a probabilistic model. Med. Image Comput. Comput. Assist. Interv. 2009, 12, 255–262. [Google Scholar]
- Sommen, F.; Zinger, S.; Schoon, E.; With, P. Supportive automatic annotation of early esophageal cancer using local gabor and color features. Neurocomputing 2014, 144, 92–106. [Google Scholar] [CrossRef]
- Yang, J.; Haas, B.; Fang, R.; Beadle, B.M.; Garden, A.S.; Liao, Z.; Zhang, L.; Balter, P.; Court, L. Atlas ranking and selection for automatic segmentation of the esophagus from CT scans. Phys. Med. Biol. 2017, 62, 9140–9158. [Google Scholar] [CrossRef]
- Mendel, R.; Ebigbo, A.; Probst, A.; Messmann, H.; Palm, C. Barrett’s esophagus analysis using convolutional neural networks. In Proceedings of the Bildverarbeitung für die Medizin 2017: Algorithmen–Systeme–Anwendungen, Proceedings des Workshops, Heidelberg, Germany, 12–14 March 2017; pp. 80–85. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Huang, G.; Zhu, J.; Li, J.; Wang, Z.; Zhou, J. Channel-attention U-Net: Channel attention mechanism for semantic segmentation of esophagus and esophageal cancer. IEEE Access 2020, 8, 122798–122810. [Google Scholar] [CrossRef]
- Tran, M.; Kim, S.; Yang, H.; Lee, G.; Oh, I.; Kang, S. Esophagus segmentation in CT images via spatial attention network and STAPLE algorithm. Sensors 2021, 21, 4456. [Google Scholar] [CrossRef]
- Chen, S.; Yang, H.; Fu, J.; Mei, W.; Ren, S.; Liu, Y.; Zhu, Z.; Liu, L.; Li, H.; Chen, H. U-Net Plus: Deep Semantic Segmentation for Esophagus and Esophageal Cancer in Computed Tomography Images. IEEE Access 2019, 7, 82867–82877. [Google Scholar] [CrossRef]
- Diniz, J.; Ferreira, J.; Diniz, P.; Silva, A.; Paova, A. Esophagus segmentation from planning CT images using an atlas-based deep learning approach. Comput. Methods Programs Biomed. 2020, 197, 105685. [Google Scholar] [CrossRef]
- Wu, Z.; Ge, R.; Wen, M.; Liu, G.; Chen, Y.; Zhang, P.; He, X.; Hua, J.; Luo, L.; Li, S. ELNet: Automatic classification and segmentation for esophageal lesions using convolutional neural network. Med. Image Anal. 2021, 67, 101838. [Google Scholar] [CrossRef]
- Chakravarty, A.; Sivswamy, J. A deep learning based joint segmentation and classification framework for glaucoma assessment in retinal color fundus images. arXiv 2018, arXiv:1808.01355. Available online: https://arxiv.org/abs/1808.01355 (accessed on 29 July 2018).
- Song, L.; Lin, J.; Wang, Z.; Wang, H. An end-to-end multi-task deep learning framework for skin lesion analysis. IEEE J. Biomed. Health Inform. 2020, 24, 2912–2921. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, H.; Du, J.; Qin, J.; Wang, T.; Chen, Y.; Liu, B.; Gao, W.; Ma, G.; Lei, B. 3D multi-attention guided multi-task learning network for automatic gastric tumor segmentation and lymph node classification. IEEE Trans. Med. Imaging 2021, 40, 1618–1631. [Google Scholar] [CrossRef]
- Lin, T.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 318–327. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.; Yang, H.; Hsiao, J.; Chen, C. Deep Learning of Binary Hash Codes for Fast Image Retrieval. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Boston, MA, USA, 11–12 June 2015; pp. 27–35. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 worlds: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. Available online: https://arxiv.org/abs/2010.11929 (accessed on 22 October 2020).
- Xie, E.; Wang, W.; Yu, Z.; Anndkumar, A.; Alvarez, J.; Luo, P. SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers. arXiv 2021, arXiv:2105.15203. Available online: http://arxiv.org/abs/2105.15203 (accessed on 31 May 2021).
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. Available online: http://arxiv.org/abs/1409.1556 (accessed on 10 April 2015).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Xie, S.; Girshick, R.; Dollar, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946v1. Available online: http://arxiv.org/abs/1905.11946v1 (accessed on 28 May 2019).
- Radosavovic, I.; Kosaraju, R.; Girshick, R.; He, K.; Dollar, P. Designing Network Design Spaces. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. arXiv 2017, arXiv:1612.01105. Available online: http://arxiv.org/abs/1612.01105 (accessed on 4 December 2016).
- Shelhamer, E.; Long, J.; Darrell, T. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 833–851. [Google Scholar]
- Huang, Z.; Wang, X.; Wei, Y.; Huang, L.; Shi, H.; Liu, W.; Huang, T. CCNet: Criss-cross attention for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Chen, X.; Wang, J. Object-contextual representations for semantic segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Glasgow, UK, 23–28 August 2020; pp. 173–190. [Google Scholar]
- Cotton, P.B. Quality endoscopists and quality endoscopy units. J. Interv. Gastroenterol. 2011, 1, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Elsken, T.; Metzen, J.H.; Hutter, F. Neural architecture search: A survey. J. Mach. Learn. Res. 2019, 20, 1–21. [Google Scholar]





| Authors | Methods | Performance |
|---|---|---|
| Münzenmayer et al. [19] | content-based image retrieval | 0.71 kappa |
| Riaz et al. [20] | autocorrelation Gabor features | 82.39% accuracy |
| Yeh et al. [21] | color coherence vector | 92.86% accuracy |
| Liu et al. [22] | support vector machines | 90.75% accuracy |
| Nakagawa et al. [12] | SSMD | 91.00% accuracy |
| Kumagai et al. [23] | GoogLeNet | 90.90% accuracy |
| Liu et al. [23] | VGGNets, etc. | 89.00% accuracy |
| Du et al. [25] | ECA-DDCNN | 90.63% accuracy |
| Igarashi et al. [26] | AlexNet | 96.50% accuracy |
| Authors | Methods | Performance |
|---|---|---|
| Sommen et al. [29] | local color and texture features | 0.95 recall |
| Yang et al. [30] | online atlas selection | 0.73 DSC |
| Mendel et al. [31] | transfer learning | 0.94 sensitivity |
| Huang et al. [33] | channel-attention U-Net | 0.725 DV |
| Tran et al. [34] | spatial attention network and STAPLE algorithm | 0.869 Dice |
| Chen et al. [35] | U-Net Plus | 0.79 DV |
| Diniz et al. [36] | Atlas-based Residual-U-Net | 0.8215 Dice |
| Models | Top-1 Accuracy ± std | F1 Score ± std |
|---|---|---|
| VGG-16 [45] | 92.68% ± 0.26 | 88.12% ± 0.26 |
| ResNet-18 [46] | 93.18% ± 0.25 | 88.36% ± 0.27 |
| ResNeXt-50 [47] | 94.34% ± 0.38 | 90.76% ± 0.33 |
| Efficientnet-B0 [48] | 95.15% ± 0.40 | 92.42% ± 0.39 |
| RegNetY-400MF [49] | 94.64% ± 0.52 | 91.57% ± 0.59 |
| Ours | 96.76% ± 0.22 | 94.22% ± 0.23 |
| Performance | Accuracy | Precision | Sensitivity | Specificity | NPV | F1-Score | |
|---|---|---|---|---|---|---|---|
| Our model | cancer | 98.48% | 98.21% | 96.49% | 99.29% | 99.59% | 97.34% |
| normal | 96.46% | 90.00% | 95.74% | 96.69% | 98.65% | 92.78% | |
| esophagitis | 95.96% | 96.74% | 94.68% | 97.12% | 95.28% | 95.70% | |
| all | 96.96% | 94.98% | 95.64% | 97.70% | 97.84% | 95.27% | |
| Endoscopists only | cancer | 91.41% | 87.04% | 82.46% | 95.04% | 93.06% | 84.69% |
| normal | 83.84% | 60.87% | 89.36% | 82.12% | 96.12% | 72.41% | |
| esophagitis | 76.26% | 81.33% | 64.89% | 86.54% | 73.17% | 72.19% | |
| all | 83.84% | 76.41% | 78.90% | 87.90% | 87.45% | 76.43% | |
| Endoscopists (single classification) | cancer | 93.43% | 95.83% | 78.90% | 98.58% | 92.67% | 87.62% |
| normal | 87.04% | 65.67% | 93.62% | 84.77% | 97.71% | 77.19% | |
| esophagitis | 81.31% | 84.34% | 74.47% | 87.5% | 79.13% | 79.10% | |
| all | 87.26% | 81.94% | 82.33% | 90.28% | 89.84% | 81.30% | |
| Endoscopists (our model) | cancer | 96.46% | 93.10% | 94.74% | 97.16% | 97.86% | 93.91% |
| normal | 90.40% | 73.33% | 93.62% | 89.4% | 97.83% | 82.24% | |
| esophagitis | 89.9% | 96.25 | 81.91% | 97.12% | 85.51% | 88.50% | |
| all | 92.25% | 87.56% | 90.09% | 94.56% | 97.73% | 88.22% | |
| Counts | Endoscopists (Before) | Total | ||
|---|---|---|---|---|
| Right | Wrong | |||
| Endoscopists (after) | Right | 152 | 27 | 175 |
| Wrong | 8 | 11 | 23 | |
| Total | 160 | 38 | 198 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Tang, S.; Cheang, C.F.; Yu, H.H.; Choi, I.C. Multi-Task Model for Esophageal Lesion Analysis Using Endoscopic Images: Classification with Image Retrieval and Segmentation with Attention. Sensors 2022, 22, 283. https://doi.org/10.3390/s22010283
Yu X, Tang S, Cheang CF, Yu HH, Choi IC. Multi-Task Model for Esophageal Lesion Analysis Using Endoscopic Images: Classification with Image Retrieval and Segmentation with Attention. Sensors. 2022; 22(1):283. https://doi.org/10.3390/s22010283
Chicago/Turabian StyleYu, Xiaoyuan, Suigu Tang, Chak Fong Cheang, Hon Ho Yu, and I Cheong Choi. 2022. "Multi-Task Model for Esophageal Lesion Analysis Using Endoscopic Images: Classification with Image Retrieval and Segmentation with Attention" Sensors 22, no. 1: 283. https://doi.org/10.3390/s22010283
APA StyleYu, X., Tang, S., Cheang, C. F., Yu, H. H., & Choi, I. C. (2022). Multi-Task Model for Esophageal Lesion Analysis Using Endoscopic Images: Classification with Image Retrieval and Segmentation with Attention. Sensors, 22(1), 283. https://doi.org/10.3390/s22010283






