Mechanistic Evaluation of Antimicrobial Lipid Interactions with Tethered Lipid Bilayers by Electrochemical Impedance Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Antimicrobial Lipid Preparation
2.3. Tethered Bilayer Lipid Membrane (tBLM) Formation
3. Results
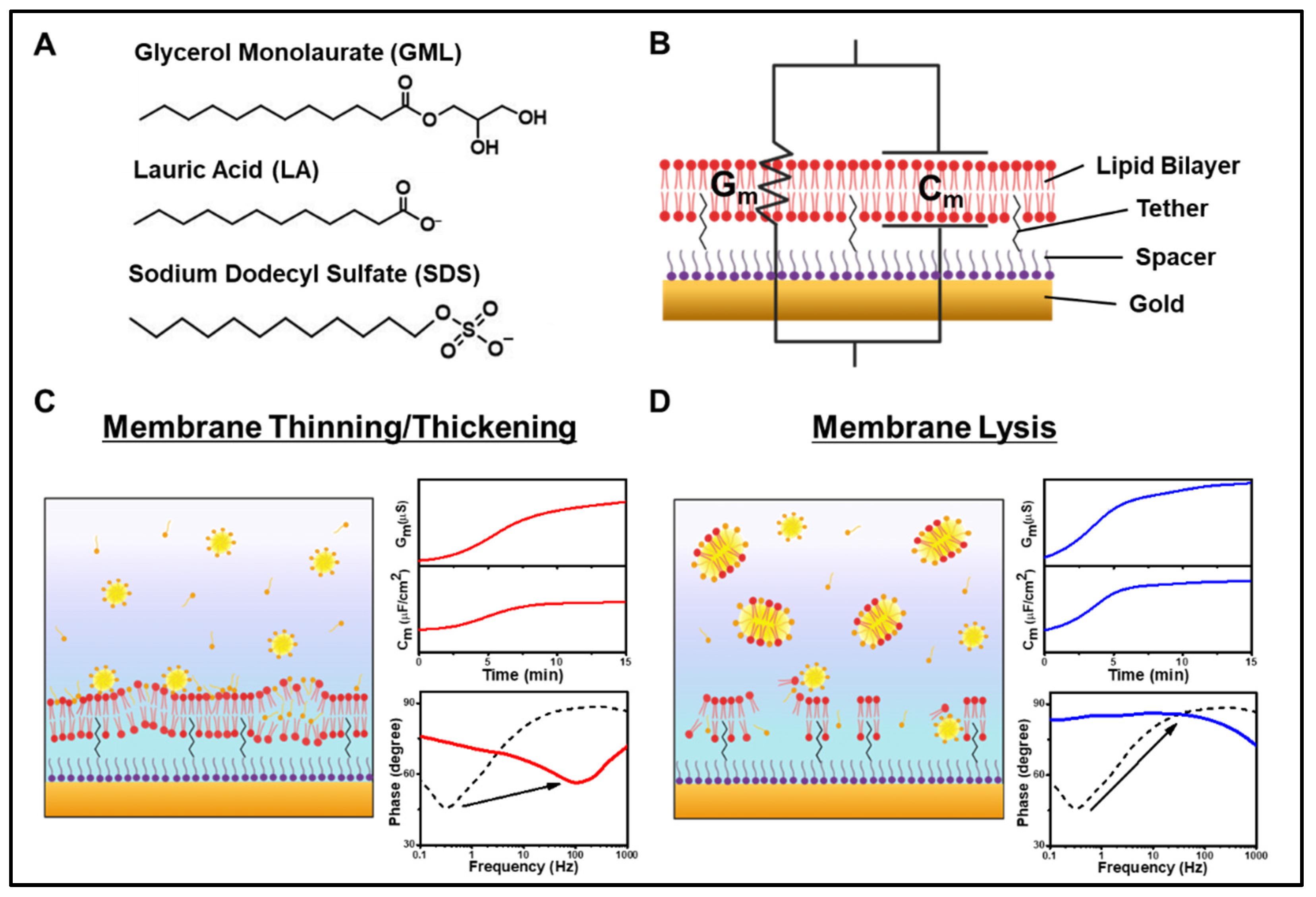
3.1. Measurement Strategy
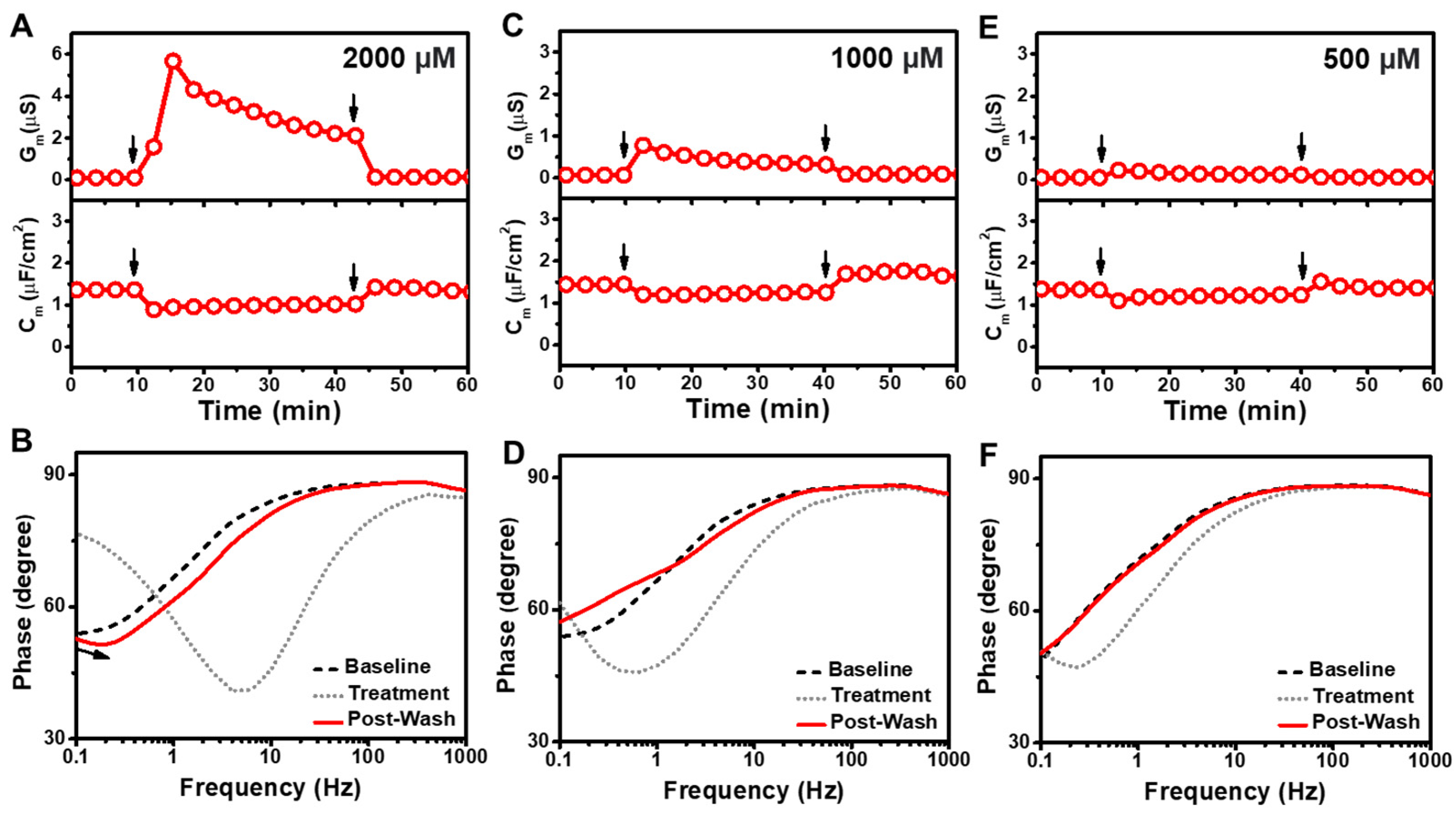
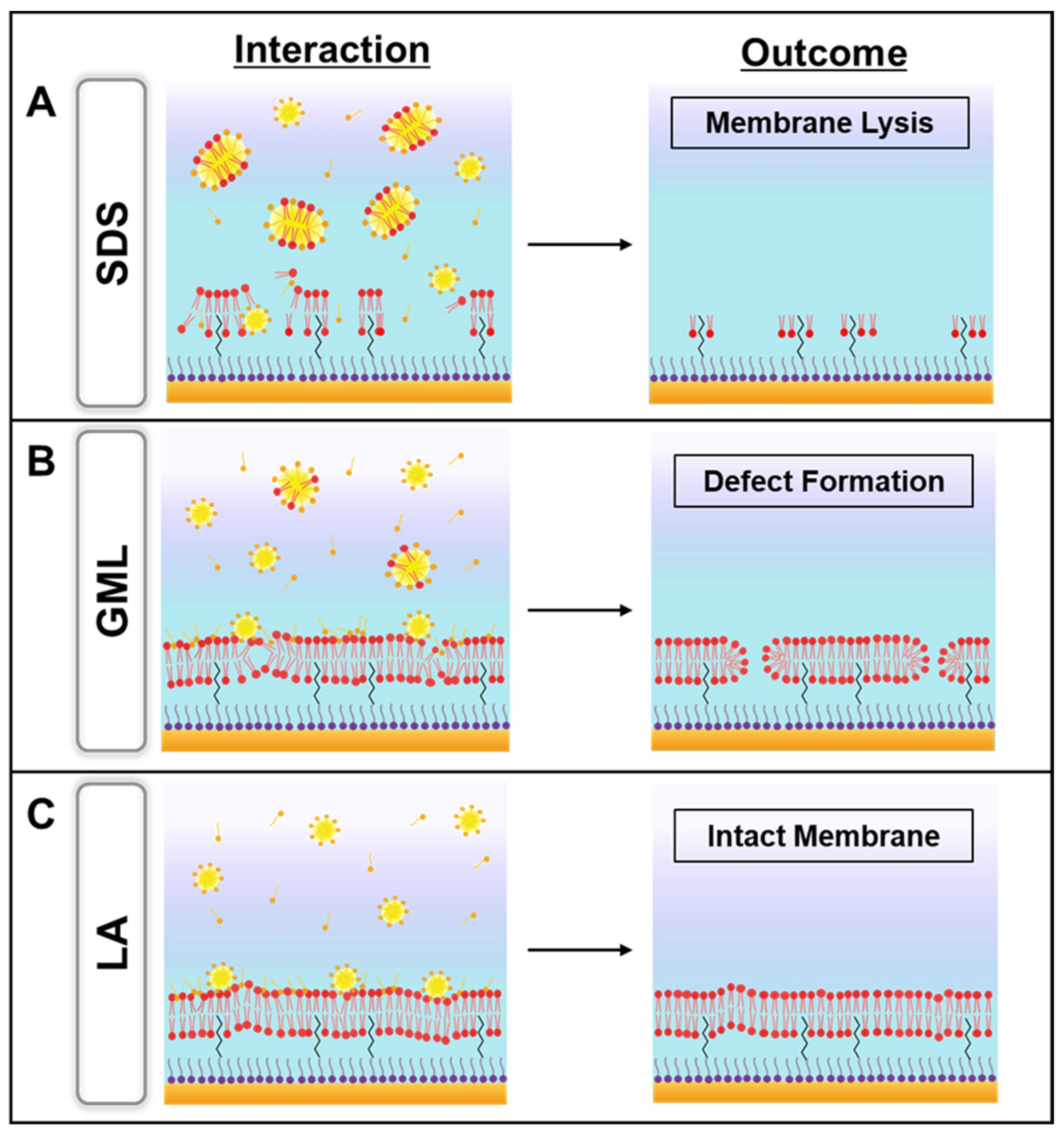
3.2. GML
3.3. LA
3.4. SDS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cheon, J.; Qin, J.; Lee, L.P.; Lee, H. Advances in Biosensor Technologies for Infection Diagnostics. Acc. Chem. Res. 2022, 55, 121–122. [Google Scholar] [CrossRef] [PubMed]
- Reder-Christ, K.; Bendas, G. Biosensor Applications in the Field of Antibiotic Research—A Review of Recent Developments. Sensors 2011, 11, 9450–9466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKeating, K.S.; Aubé, A.; Masson, J.-F. Biosensors and Nanobiosensors for Therapeutic Drug and Response Monitoring. Analyst 2016, 141, 429–449. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.A.A.; Rahman, S.; Cohall, D.; Bharatha, A.; Singh, K.; Haque, M.; Gittens-St Hilaire, M. Antimicrobial Stewardship: Fighting Antimicrobial Resistance and Protecting Global Public Health. Infect. Drug Resist. 2020, 13, 4713–4738. [Google Scholar] [CrossRef]
- Zucca, M.; Savoia, D. The Post-Antibiotic Era: Promising Developments in the Therapy of Infectious Diseases. Int. J. Biomed. Sci. 2010, 6, 77–86. [Google Scholar]
- Kumar, M.; Sarma, D.K.; Shubham, S.; Kumawat, M.; Verma, V.; Nina, P.B.; Jp, D.; Kumar, S.; Singh, B.; Tiwari, R.R. Futuristic Non-Antibiotic Therapies to Combat Antibiotic Resistance: A Review. Front. Microbiol. 2021, 12, 609459. [Google Scholar] [CrossRef]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef]
- Borrelli, L.; Varriale, L.; Dipineto, L.; Pace, A.; Menna, L.F.; Fioretti, A. Insect Derived Lauric Acid as Promising Alternative Strategy to Antibiotics in the Antimicrobial Resistance Scenario. Front. Microbiol. 2021, 12, 620798. [Google Scholar] [CrossRef]
- Alves, E.; Dias, M.; Lopes, D.; Almeida, A.; Domingues, M.D.; Rey, F. Antimicrobial Lipids from Plants and Marine Organisms: An Overview of the Current State-of-the-Art and Future Prospects. Antibiotics 2020, 9, 441. [Google Scholar] [CrossRef]
- Yoon, B.K.; Jackman, J.A.; Kim, M.C.; Cho, N.-J. Spectrum of Membrane Morphological Responses to Antibacterial Fatty Acids and Related Surfactants. Langmuir 2015, 31, 10223–10232. [Google Scholar] [CrossRef]
- Valle-González, E.R.; Jackman, J.A.; Yoon, B.K.; Park, S.; Sut, T.N.; Cho, N.-J. Characterizing How Acidic pH Conditions Affect the Membrane-Disruptive Activities of Lauric Acid and Glycerol Monolaurate. Langmuir 2018, 34, 13745–13753. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, C.; Hu, X.; Yao, X.; Han, L.; Wu, X.; Li, R.; Wen, T.; Ming, L. Lauric Acid Induces Apoptosis of Rice Sheath Blight Disease Caused by Rhizoctonia solani by Affecting Fungal Fatty Acid Metabolism and Destroying the Dynamic Equilibrium of Reactive Oxygen Species. J. Fungi 2022, 8, 153. [Google Scholar] [CrossRef] [PubMed]
- Matsue, M.; Mori, Y.; Nagase, S.; Sugiyama, Y.; Hirano, R.; Ogai, K.; Ogura, K.; Kurihara, S.; Okamoto, S. Measuring the Antimicrobial Activity of Lauric Acid against Various Bacteria in Human Gut Microbiota Using a New Method. Cell Transplant. 2019, 28, 1528–1541. [Google Scholar] [CrossRef] [PubMed]
- Schlievert, P.M.; Kilgore, S.H.; Seo, K.S.; Leung, D.Y.M. Glycerol Monolaurate Contributes to the Antimicrobial and Anti-Inflammatory Activity of Human Milk. Sci. Rep. 2019, 9, 14550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thormar, H.; Isaacs, C.; Brown, H.R.; Barshatzky, M.R.; Pessolano, T. Inactivation of Enveloped Viruses and Killing of Cells by Fatty Acids and Monoglycerides. Antimicrob. Agents Chemother. 1987, 31, 27–31. [Google Scholar] [CrossRef] [Green Version]
- Jackman, J.A.; Hakobyan, A.; Zakaryan, H.; Elrod, C.C. Inhibition of African Swine Fever Virus in Liquid and Feed by Medium-Chain Fatty Acids and Glycerol Monolaurate. J. Anim. Sci. Biotechnol. 2020, 11, 114. [Google Scholar] [CrossRef]
- Parsons, J.B.; Yao, J.; Frank, M.W.; Jackson, P.; Rock, C.O. Membrane Disruption by Antimicrobial Fatty Acids Releases Low-Molecular-Weight Proteins from Staphylococcus aureus. J. Bacteriol. 2012, 194, 5294–5304. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.-T.; Chen, J.-W.; Rathod, J.; Jiang, Y.-Z.; Tsai, P.-J.; Hung, Y.-P.; Ko, W.-C.; Paredes-Sabja, D.; Huang, I.-H. Lauric Acid Is an Inhibitor of Clostridium difficile Growth in Vitro and Reduces Inflammation in a Mouse Infection Model. Front. Microbiol. 2018, 8, 2635. [Google Scholar] [CrossRef] [Green Version]
- Osaki, T.; Takeuchi, S. Artificial Cell Membrane Systems for Biosensing Applications. Anal. Chem. 2017, 89, 216–231. [Google Scholar] [CrossRef]
- Reimhult, E.; Kumar, K. Membrane Biosensor Platforms Using Nano- and Microporous Supports. Trends Biotechnol. 2008, 26, 82–89. [Google Scholar] [CrossRef]
- Hyldgaard, M.; Sutherland, D.; Sundh, M.; Mygind, T.; Meyer, R. Antimicrobial Mechanism of Monocaprylate. Appl. Environ. Microbiol. 2012, 78, 2957–2965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, B.K.; Park, H.; Zhdanov, V.P.; Jackman, J.A.; Cho, N.-J. Real-Time Nanoplasmonic Sensing of Three-Dimensional Morphological Changes in a Supported Lipid Bilayer and Antimicrobial Testing Applications. Biosens. Bioelectron. 2021, 174, 112768. [Google Scholar] [CrossRef] [PubMed]
- Yoon, B.K.; Jackman, J.A.; Kim, M.C.; Sut, T.N.; Cho, N.J. Correlating Membrane Morphological Responses with Micellar Aggregation Behavior of Capric Acid and Monocaprin. Langmuir 2017, 33, 2750–2759. [Google Scholar] [CrossRef] [PubMed]
- Yoon, B.K.; Jackman, J.A.; Valle-Gonzalez, E.R.; Cho, N.J. Antibacterial Free Fatty Acids and Monoglycerides: Biological Activities, Experimental Testing, and Therapeutic Applications. Int. J. Mol. Sci. 2018, 19, 1114. [Google Scholar] [CrossRef] [Green Version]
- Briand, E.; Zäch, M.; Svedhem, S.; Kasemo, B.; Petronis, S. Combined QCM-D and EIS Study of Supported Lipid Bilayer Formation and Interaction with Pore-Forming Peptides. Analyst 2010, 135, 343–350. [Google Scholar] [CrossRef]
- Andersson, J.; Köper, I.; Knoll, W. Tethered Membrane Architectures—Design and Applications. Front. Mater. 2018, 5, 55. [Google Scholar] [CrossRef] [Green Version]
- Junghans, A.; Köper, I. Structural Analysis of Tethered Bilayer Lipid Membranes. Langmuir 2010, 26, 11035–11040. [Google Scholar] [CrossRef]
- Hoiles, W.; Gupta, R.; Cornell, B.; Cranfield, C.; Krishnamurthy, V. The Effect of Tethers on Artificial Cell Membranes: A Coarse-Grained Molecular Dynamics Study. PLoS ONE 2016, 11, e0162790. [Google Scholar] [CrossRef]
- Liu, C.; Faller, R. Conformational, Dynamical. and Tensional Study of Tethered Bilayer Lipid Membranes in Coarse-Grained Molecular Simulations. Langmuir 2012, 28, 15907–15915. [Google Scholar] [CrossRef]
- Jackman, J.A.; Knoll, W.; Cho, N.-J. Biotechnology Applications of Tethered Lipid Bilayer Membranes. Materials 2012, 5, 2637–2657. [Google Scholar] [CrossRef] [Green Version]
- Vockenroth, I.K.; Ohm, C.; Robertson, J.W.F.; McGillivray, D.J.; Lösche, M.; Köper, I. Stable Insulating Tethered Bilayer Lipid Membranes. Biointerphases 2008, 3, FA68–FA73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cranfield, C.G.; Cornell, B.A.; Grage, S.L.; Duckworth, P.; Carne, S.; Ulrich, A.S.; Martinac, B. Transient Potential Gradients and Impedance Measures of Tethered Bilayer Lipid Membranes: Pore-Forming Peptide Insertion and the Effect of Electroporation. Biophys. J. 2014, 106, 182–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alghalayini, A.; Garcia, A.; Berry, T.; Cranfield, C.G. The Use of Tethered Bilayer Lipid Membranes to Identify the Mechanisms of Antimicrobial Peptide Interactions with Lipid Bilayers. Antibiotics 2019, 8, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Jackman, J.A.; Cho, N.-J. Comparing the Membrane-Interaction Profiles of Two Antiviral Peptides: Insights into Structure–Function Relationship. Langmuir 2019, 35, 9934–9943. [Google Scholar] [CrossRef] [PubMed]
- Cranfield, C.G.; Berry, T.; Holt, S.A.; Hossain, K.R.; Le Brun, A.P.; Carne, S.; Al Khamici, H.; Coster, H.; Valenzuela, S.M.; Cornell, B. Evidence of the Key Role of H3O+ in Phospholipid Membrane Morphology. Langmuir 2016, 32, 10725–10734. [Google Scholar] [CrossRef] [PubMed]
- Berry, T.; Dutta, D.; Chen, R.; Leong, A.; Wang, H.; Donald, W.A.; Parviz, M.; Cornell, B.; Willcox, M.; Kumar, N.; et al. Lipid Membrane Interactions of the Cationic Antimicrobial Peptide Chimeras Melimine and Cys-Melimine. Langmuir 2018, 34, 11586–11592. [Google Scholar] [CrossRef]
- Alghalayini, A.; Jiang, L.; Gu, X.; Yeoh, G.H.; Cranfield, C.G.; Timchenko, V.; Cornell, B.A.; Valenzuela, S.M. Real-Time Monitoring of Heat Transfer between Gold Nanoparticles and Tethered Bilayer Lipid Membranes. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183334. [Google Scholar] [CrossRef]
- Alghalayini, A.; Jiang, L.; Gu, X.; Yeoh, G.H.; Cranfield, C.G.; Timchenko, V.; Cornell, B.A.; Valenzuela, S.M. Tethered Bilayer Lipid Membranes to Monitor Heat Transfer between Gold Nanoparticles and Lipid Membranes. J. Vis. Exp. 2020, 166, e61851. [Google Scholar] [CrossRef]
- Alharbi, A.R.M.; Andersson, J.M.; Köper, I.; Andersson, G.G. Investigating the Structure of Self-Assembled Monolayers Related to Biological Cell Membranes. Langmuir 2019, 35, 14213–14221. [Google Scholar] [CrossRef]
- Cranfield, C.; Carne, S.; Martinac, B.; Cornell, B. The Assembly and Use of Tethered Bilayer Lipid Membranes (tBLMs). Methods Mol. Biol. 2015, 1232, 45–53. [Google Scholar]
- Cseresnyés, I.; Rajkai, K.; Vozáry, E. Role of Phase Angle Measurement in Electrical Impedance Spectroscopy. Int. Agrophys. 2013, 27, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Cranfield, C.G.; Henriques, S.T.; Martinac, B.; Duckworth, P.; Craik, D.J.; Cornell, B. Kalata B1 and Kalata B2 Have a Surfactant-Like Activity in Phosphatidylethanolomine-Containing Lipid Membranes. Langmuir 2017, 33, 6630–6637. [Google Scholar] [CrossRef] [PubMed]
- Simard, J.R.; Kamp, F.; Hamilton, J.A. Measuring the Adsorption of Fatty Acids to Phospholipid Vesicles by Multiple Fluorescence Probes. Biophys. J. 2008, 94, 4493–4503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.; Lu, X.; Deng, Z.; Xiao, S.; Yuan, B.; Yang, K. How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane. Molecules 2019, 24, 1775. [Google Scholar] [CrossRef] [Green Version]
- Salditt, T.; Li, C.; Spaar, A. Structure of Antimicrobial Peptides and Lipid Membranes Probed by Interface-Sensitive X-ray Scattering. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1483–1498. [Google Scholar] [CrossRef]
- Pachioni, J.; Magalhães, J.; Lima, E.; De Moura Bueno, L.; Barbosa, J.; Malta de Sá, M.; Rangel-Yagui, C. Alkylphospholipids–A Promising Class of Chemotherapeutic Agents with a Broad Pharmacological Spectrum. J. Pharm. Pharm. Sci. 2013, 16, 742–759. [Google Scholar] [CrossRef] [Green Version]
- Fosdick, M.G.; Chheda, P.R.; Tran, P.M.; Wolff, A.; Peralta, R.; Zhang, M.Y.; Kerns, R.; Houtman, J.C.D. Suppression of Human T Cell Activation by Derivatives of Glycerol Monolaurate. Sci. Rep. 2021, 11, 8943. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Peterson, M.L. Glycerol Monolaurate Antibacterial Activity in Broth and Biofilm Cultures. PLoS ONE 2012, 7, e40350. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.S.; Sandouk, A.; Houtman, J.C.D. Glycerol Monolaurate (GML) Inhibits Human T Cell Signaling and Function by Disrupting Lipid Dynamics. Sci. Rep. 2016, 6, 30225. [Google Scholar] [CrossRef] [Green Version]
- Pouny, Y.; Rapaport, D.; Mor, A.; Nicolas, P.; Shai, Y. Interaction of Antimicrobial Dermaseptin and its Fluorescently Labeled Analogs with Phospholipid Membranes. Biochemistry 1992, 31, 12416–12423. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Huster, D.; Waring, A.; Lehrer, R.I.; Kearney, W.; Tack, B.F.; Hong, M. Orientation and Dynamics of an Antimicrobial Peptide in the Lipid Bilayer by Solid-State NMR Spectroscopy. Biophys. J. 2001, 81, 2203–2214. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.; Yoon, B.K.; Jackman, J.A. Effect of Membrane Curvature Nanoarchitectonics on Membrane-Disruptive Interactions of Antimicrobial Lipids and Surfactants. Langmuir 2022, 38, 4606–4616. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, S.W.; Jeon, W.-Y.; Yoon, B.K.; Jackman, J.A. Mechanistic Evaluation of Antimicrobial Lipid Interactions with Tethered Lipid Bilayers by Electrochemical Impedance Spectroscopy. Sensors 2022, 22, 3712. https://doi.org/10.3390/s22103712
Tan SW, Jeon W-Y, Yoon BK, Jackman JA. Mechanistic Evaluation of Antimicrobial Lipid Interactions with Tethered Lipid Bilayers by Electrochemical Impedance Spectroscopy. Sensors. 2022; 22(10):3712. https://doi.org/10.3390/s22103712
Chicago/Turabian StyleTan, Sue Woon, Won-Yong Jeon, Bo Kyeong Yoon, and Joshua A. Jackman. 2022. "Mechanistic Evaluation of Antimicrobial Lipid Interactions with Tethered Lipid Bilayers by Electrochemical Impedance Spectroscopy" Sensors 22, no. 10: 3712. https://doi.org/10.3390/s22103712






