Histopathologic Oral Cancer Prediction Using Oral Squamous Cell Carcinoma Biopsy Empowered with Transfer Learning
Abstract
:1. Introduction
2. Literature Review
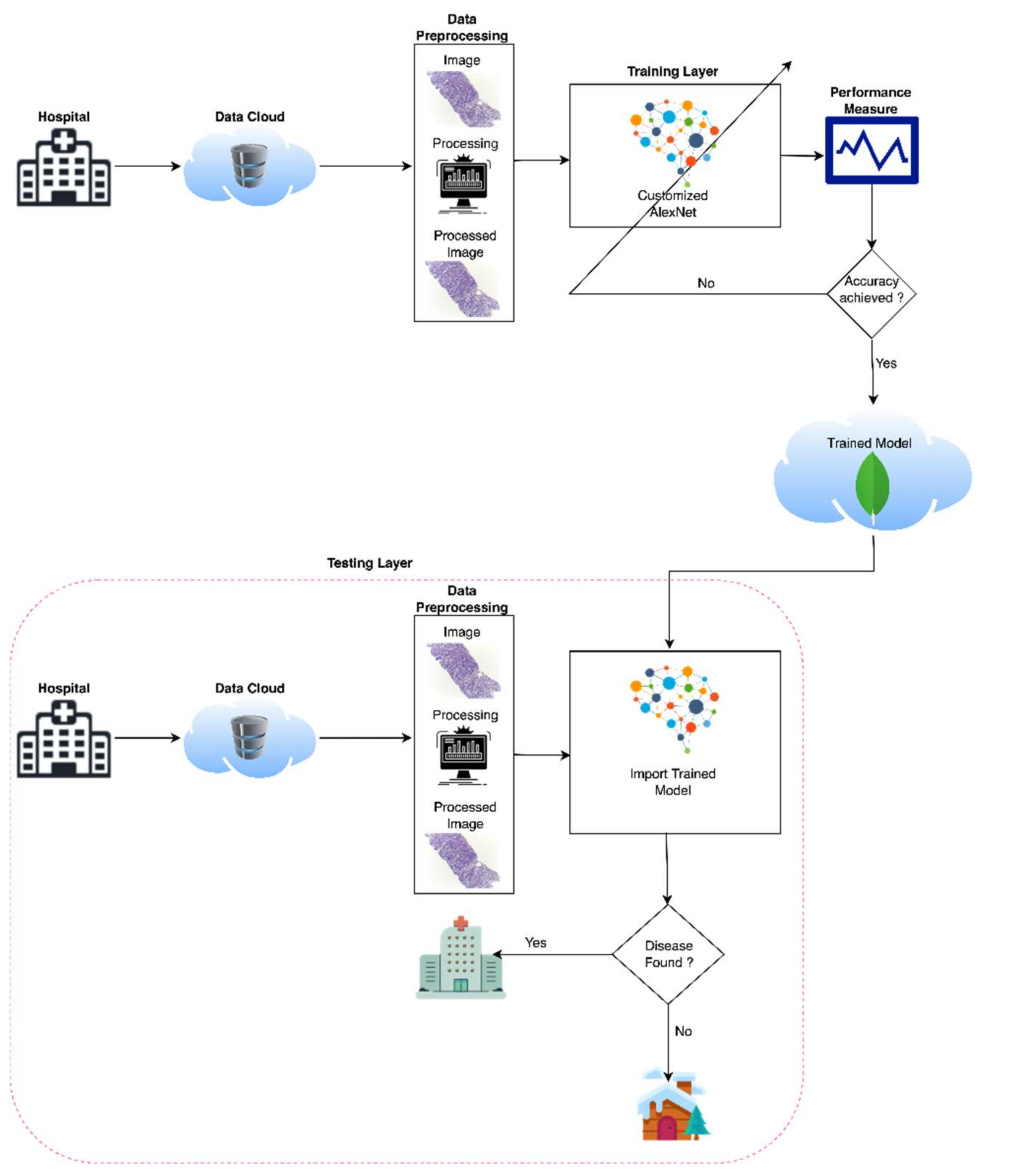
3. Materials and Methods
4. Data Set
5. Customized AlexNet
6. Simulation and Results
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. WHO Report on Cancer: Setting Priorities, Investing Wisely and Providing Care for All; Technical Report; World Health Organization: Geneva, Switzerland, 2020.
- Sinevici, N.; O’sullivan, J. Oral cancer: Deregulated molecular events and their use as biomarkers. Oral Oncol. 2016, 61, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Ilhan, B.; Lin, K.; Guneri, P.; Wilder-Smith, P. Improving Oral Cancer Outcomes with Imaging and Artificial Intelligence. J. Dent. Res. 2020, 99, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Dhanuthai, K.; Rojanawatsirivej, S.; Thosaporn, W.; Kintarak, S.; Subarnbhesaj, A.; Darling, M.; Kryshtalskyj, E.; Chiang, C.P.; Shin, H.I. Oral cancer: A multicenter study. Med. Oral Patol. Oral Cir. Bucal 2018, 23, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Lavanya, L.; Chandra, J. Oral Cancer Analysis Using Machine Learning Techniques. Int. J. Eng. Res. Technol. 2019, 12, 596–601. [Google Scholar]
- Kearney, V.; Chan, J.W.; Valdes, G.; Solberg, T.D.; Yom, S.S. The application of artificial intelligence in the IMRT planning process for head and neck cancer. Oral Oncol. 2018, 87, 111–116. [Google Scholar] [CrossRef]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69, 36–40. [Google Scholar] [CrossRef]
- Head and Neck sq Squamous Cell Carcinoma. Available online: https://en.wikipedia.org/wiki/Head_and_neck_sqsquamous-cell_carcinoma (accessed on 12 January 2022).
- Kaladhar, D.; Chandana, B.; Kumar, P. Predicting Cancer Survivability Using Classification Algorithms. Int. J. Res. Rev. Comput. Sci. 2011, 2, 340–343. [Google Scholar]
- Kalappanavar, A.; Sneha, S.; Annigeri, R.G. Artificial intelligence: A dentist’s perspective. J. Med. Radiol. Pathol. Surg. 2018, 5, 2–4. [Google Scholar] [CrossRef]
- Krishna, A.B.; Tanveer, A.; Bhagirath, P.V.; Gannepalli, A. Role of artificial intelligence in diagnostic oral pathology-A modern approach. J. Oral Maxillofac. Pathol. JOMFP 2020, 24, 152–156. [Google Scholar] [CrossRef]
- Kareem, S.A.; Pozos-Parra, P.; Wilson, N. An application of belief merging for the diagnosis of oral cancer. Appl. Soft Comput. J. 2017, 61, 1105–1112. [Google Scholar] [CrossRef]
- Kann, B.H.; Aneja, S.; Loganadane, G.V.; Kelly, J.R.; Smith, S.M.; Decker, R.H.; Yu, J.B. Pretreatment Identification of Head and Neck Cancer Nodal Metastasis and Extranodal Extension Using Deep Learning Neural Networks. Sci. Rep. 2018, 8, 14306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohd, F.; Noor, N.M.; Abu Bakar, Z.; Rajion, Z.A. Analysis of Oral Cancer Prediction using Features Selection with Machine Learning. In Proceedings of the ICIT The 7th International Conference on Information Technology, Amman, Jordan, 12–15 May 2015. [Google Scholar]
- Ahmad, L.G.; Eshlaghy, A.G.; Poorebrahimi, A.; Ebrahimi, M.; Razavi, A.R. Using Three Machine Learning Techniques for Predicting Breast Cancer Recurrence. Health Med. Inform. 2013, 4, 2157–7420. [Google Scholar]
- Vidhya, M.S.; Nisha, D.S.S.; Sathik, D.M.M. Denoising the CT Images for Oropharyngeal Cancer using Filtering Techniques. Int. J. Eng. Res. Technol. 2020, 8, 1–4. [Google Scholar]
- Chakraborty, D.; Natarajan, C.; Mukherjee, A. Advances in oral cancer detection. Adv. Clin. Chem. 2019, 91, 181–200. [Google Scholar]
- Anitha, N.; Jamberi, K. Diagnosis, and Prognosis of Oral Cancer using classification algorithm with Data Mining Techniques. Int. J. Data Min. Tech. Appl. 2017, 6, 59–61. [Google Scholar]
- Available online: https://www.ahns.info/resources/education/patient_education/oralcavity (accessed on 12 January 2022).
- Aubreville, M.; Knipfer, C.; Oetter, N.; Jaremenko, C.; Rodner, E.; Denzler, J.; Bohr, C.; Neumann, H.; Stelzle, F.; Maier, A. Automatic Classification of Cancerous Tissue in Laserendomicroscopy Images of the Oral Cavity using Deep Learning. Sci. Rep. 2017, 7, 11979. [Google Scholar] [CrossRef] [Green Version]
- Alabi, R.O.; Youssef, O.; Pirinen, M.; Elmusrati, M.; Mäkitie, A.A.; Leivo, I.; Almangush, A. Machine learning in oral squamous cell carcinoma: Current status, clinical concerns and prospects for future—A systematic review. Artif. Intell. Med. 2021, 115, 102060. [Google Scholar] [CrossRef]
- Sun, M.-L.; Liu, Y.; Liu, G.-M.; Cui, D.; Heidari, A.A.; Jia, W.-Y.; Ji, X.; Chen, H.-L.; Luo, Y.-G. Application of Machine Learning to Stomatology: A Comprehensive Review. IEEE Access 2020, 8, 184360–184374. [Google Scholar] [CrossRef]
- Alkhadar, H.; Macluskey, M.; White, S.; Ellis, I.; Gardner, A. Comparison of machine learning algorithms for the prediction of five-year survival in oral squamous cell carcinoma. J. Oral Pathol. Med. 2021, 50, 378–384. [Google Scholar] [CrossRef]
- Saba, T. Recent advancement in cancer detection using machine learning: Systematic survey of decades, comparisons and challenges. J. Infect. Public Health 2020, 13, 1274–1289. [Google Scholar] [CrossRef]
- Kouznetsova, V.L.; Li, J.; Romm, E.; Tsigelny, I.F. Finding distinctions between oral cancer and periodontitis using saliva metabolites and machine learning. Oral Dis. 2021, 27, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Alhazmi, A.; Alhazmi, Y.; Makrami, A.; Salawi, N.; Masmali, K.; Patil, S. Application of artificial intelligence and machine learning for prediction of oral cancer risk. J. Oral Pathol. Med. 2021, 50, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.S.; Lee, N.P.; Adeoye, J.; Thomson, P.; Choi, S.W. Machine learning and treatment outcome prediction for oral cancer. J. Oral Pathol. Med. 2020, 49, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Welikala, R.A.; Remagnino, P.; Lim, J.H.; Chan, C.S.; Rajendran, S.; Kallarakkal, T.G.; Zain, R.B.; Jayasinghe, R.D.; Rimal, J.; Kerr, A.R.; et al. Automated Detection and Classification of Oral Lesions Using Deep Learning for Early Detection of Oral Cancer. IEEE Access 2020, 8, 132677–132693. [Google Scholar] [CrossRef]
- Mehmood, S.; Ghazal, T.M.; Khan, M.A.; Zubair, M.; Naseem, M.T.; Faiz, T.; Ahmad, M. Malignancy Detection in Lung and Colon Histopathology Images Using Transfer Learning with Class Selective Image Processing. IEEE Access 2022, 10, 25657–25668. [Google Scholar] [CrossRef]
- Ahmed, U.; Issa, G.F.; Khan, M.A.; Aftab, S.; Said, R.A.T.; Ghazal, T.M.; Ahmad, M. Prediction of Diabetes Empowered with Fused Machine Learning. IEEE Access 2022, 10, 8529–8538. [Google Scholar] [CrossRef]
- Siddiqui, S.Y.; Haider, A.; Ghazal, T.M.; Khan, M.A.; Naseer, I.; Abbas, S.; Rahman, M.; Khan, J.A.; Ahmad, M.; Hasan, M.K.; et al. IoMT Cloud-Based Intelligent Prediction of Breast Cancer Stages Empowered with Deep Learning. IEEE Access 2021, 9, 146478–146491. [Google Scholar] [CrossRef]
- Sikandar, M.; Sohail, R.; Saeed, Y.; Zeb, A.; Zareei, M.; Khan, M.A.; Khan, A.; Aldosary, A.; Mohamed, E.M. Analysis for Disease Gene Association Using Machine Learning. IEEE Access 2020, 8, 160616–160626. [Google Scholar] [CrossRef]
- Kuntz, S.; Krieghoff-Henning, E.; Kather, J.N.; Jutzi, T.; Höhn, J.; Kiehl, L.; Hekler, A.; Alwers, E. Gastrointestinal cancer classification and prognostication from histology using deep learning: Systematic review. Eur. J. Cancer 2021, 155, 200–215. [Google Scholar] [CrossRef]
- Heuvelmans, M.A.; Ooijen, P.M.A.; Sarim, A.; Silva, C.F.; Han, D.; Heussel, C.P.; Hickes, W.; Kauczor, H.-U.; Novotny, P.; Peschl, H.; et al. Lung cancer prediction by Deep Learning to identify benign lung nodules. Lung Cancer 2021, 154, 1–4. [Google Scholar] [CrossRef]
- Ibrahim, D.M.; Elshennawy, N.M.; Sarhan, A.M. Deep-chest: Multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Comput. Biol. Med. 2021, 132, 104348. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, D.; Liu, Z.; Li, Z.; Xie, P.; Sun, K.; Wei, W.; Dai, W.; Tang, Z. Deep learning radiomics-based prediction of distant metastasis in patients with locally advanced rectal cancer after neoadjuvant chemoradiotherapy: A multicentre study. EBio Med. 2021, 69, 103442. [Google Scholar] [CrossRef] [PubMed]
- Wafaa Shams, K.; Zaw, Z. Oral Cancer Prediction Using Gene Expression Profiling and Machine Learning. Int. J. Appl. Eng. Res. 2017, 12, 0973–4562. [Google Scholar]
- Kalantari, N.; Ramamoorthi, R. Deep high dynamic range imaging of dynamic scenes. ACM Trans. Graph. 2017, 36, 144:1–144:12. [Google Scholar] [CrossRef]
- Deepak Kumar, J.; Surendra Bilouhan, D.; Kumar, R.C. An approach for hyperspectral image classification by optimizing SVM using self-organizing map. J. Comput. Sci. 2018, 25, 252–259. [Google Scholar]
- Shavlokhova, V.; Sandhu, S.; Flechtenmacher, C.; Koveshazi, I.; Neumeier, F.; Padrón-Laso, V.; Jonke, Ž.; Saravi, B.; Vollmer, M.; Vollmer, A.; et al. Deep Learning on Oral Squamous Cell Carcinoma Ex Vivo Fluorescent Confocal Microscopy Data: A Feasibility Study. J. Clin. Med. 2021, 10, 5326. [Google Scholar] [CrossRef]
- Available online: https://www.kaggle.com/ashenafifasilkebede/dataset?select=val (accessed on 6 January 2022).
- Chen, C.; Zhou, K.; Zha, M.; Qu, X.; Guo, X.; Chen, H.; Xiao, R. An effective deep neural network for lung lesions segmentation from COVID-19 CT images. IEEE Trans. Ind. Inform. 2021, 17, 6528–6538. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; Chang, V.; Mohamed, R. HSMA_WOA: A hybrid novel Slime mould algorithm with whale optimization algorithm for tackling the image segmentation problem of chest X-ray images. Appl. Soft Comput. 2020, 95, 106642. [Google Scholar] [CrossRef]
- Rahman, A.-U.; Abbas, S.; Gollapalli, M.; Ahmed, R.; Aftab, S.; Ahmad, M.; Khan, M.A.; Mosavi, A. Rainfall Prediction System Using Machine Learning Fusion for Smart Cities. Sensors 2022, 22, 3504. [Google Scholar] [CrossRef]
- Saleem, M.; Abbas, S.; Ghazal, T.M.; Khan, M.A.; Sahawneh, N.; Ahmad, M. Smart cities: Fusion-based intelligent traffic congestion control system for vehicular networks using machine learning techniques. Egypt. Inform. J. 2022; in press. [Google Scholar] [CrossRef]
- Nadeem, M.W.; Goh, H.G.; Khan, M.A.; Hussain, M.; Mushtaq, M.F.; Ponnusamy, V.A. Fusion-based machine learning architecture for heart disease prediction. Comput. Mater. Contin. 2021, 67, 2481–2496. [Google Scholar] [CrossRef]
- Siddiqui, S.Y.; Athar, A.; Khan, M.A.; Abbas, S.; Saeed, Y.; Khan, M.F.; Hussain, M. Modelling, simulation and optimization of diagnosis cardiovascular disease using computational intelligence approaches. J. Med. Imaging Health Inform. 2020, 10, 1005–1022. [Google Scholar] [CrossRef]






| Publication | Method | Dataset | Accuracy | Limitation |
|---|---|---|---|---|
| A.Alhazmi [26] | ANN | Public | 78.95% | Requires data preprocessing |
| C.S. Chu [27] | SVM, KNN | Public | 70.59% | Requires data preprocessing |
| R.A.Welikala [28] | ResNet101 | Public | 78.30% | Requires data preprocessing and learning criteria decision method |
| V. Shavlokhova [40] | CNN | Private | 77.89% | Requires better image data preprocessing techniques and learning criteria method |
| M. Aberville [20] | Deep Learning | Public | 80.01% | Requires data image preprocessing techniquesClass instances |
| H. Alkhadar [23] | KNN, Logistic Regression, Decision Tree, Random Forest | Public | 76% | Requires handcrafted features |
| 1 | Start |
| 2 | Input Oral Cancer Data from Data Cloud |
| 3 | Pre-process Oral Cancer data |
| 4 | Load Data |
| 5 | Load Customized Model |
| 6 | Prediction of Oral Cancer using Transfer Learning (AlexNet) |
| 7 | Training Phase |
| 8 | Image Testing Phase |
| 9 | Compute the Performance and Accuracy of the proposed model by using the Performance Matrix |
| 10 | Finish |
| Classes | No. of Images |
|---|---|
| Sick (OSCC) | 2511 |
| Healthy | 2435 |
| No. of Epochs | Learning Rate | No. of Layers | Image Dimension | Pooling Method | Mini-Batch Loss |
|---|---|---|---|---|---|
| 10 | 0.001 | 25 | 227 × 227 × 3 | MAX | 2.5674 |
| 20 | 2.3498 | ||||
| 30 | 1.3600 | ||||
| 40 | 1.4948 | ||||
| 50 | 6.1029 | ||||
| 60 | 0.2491 | ||||
| 70 | 0.3736 |
| No. of Epochs | Learning Rate | Accuracy (%) | Loss Rate (%) | Iterations | Time Elapsed (hh:mm:ss) |
|---|---|---|---|---|---|
| 10 | 0.001 | 76.12 | 23.88 | 38 per epoch | 00:03:15 |
| 20 | 80.35 | 19.65 | 00:03:45 | ||
| 30 | 86.15 | 13.85 | 00:04:34 | ||
| 40 | 90.62 | 9.38 | 00:04:55 | ||
| 50 | 85.94 | 14.06 | 00:06:11 | ||
| 60 | 94.44 | 5.56 | 00:07:17 | ||
| 70 | 97.66 | 2.34 | 00:08:34 |
| Instances (1483) | Testing (%) |
|---|---|
| CA | 90.02 |
| CMR | 9.08 |
| Sensitivity | 92.74 |
| Specificity | 87.38 |
| F1-Score | 90.15 |
| PPV | 87.69 |
| NPV | 92.55 |
| FPR | 12.62 |
| FNR | 7.26 |
| LPR | 7.35 |
| LNR | 0.08 |
| FMI | 90.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, A.-u.; Alqahtani, A.; Aldhafferi, N.; Nasir, M.U.; Khan, M.F.; Khan, M.A.; Mosavi, A. Histopathologic Oral Cancer Prediction Using Oral Squamous Cell Carcinoma Biopsy Empowered with Transfer Learning. Sensors 2022, 22, 3833. https://doi.org/10.3390/s22103833
Rahman A-u, Alqahtani A, Aldhafferi N, Nasir MU, Khan MF, Khan MA, Mosavi A. Histopathologic Oral Cancer Prediction Using Oral Squamous Cell Carcinoma Biopsy Empowered with Transfer Learning. Sensors. 2022; 22(10):3833. https://doi.org/10.3390/s22103833
Chicago/Turabian StyleRahman, Atta-ur, Abdullah Alqahtani, Nahier Aldhafferi, Muhammad Umar Nasir, Muhammad Farhan Khan, Muhammad Adnan Khan, and Amir Mosavi. 2022. "Histopathologic Oral Cancer Prediction Using Oral Squamous Cell Carcinoma Biopsy Empowered with Transfer Learning" Sensors 22, no. 10: 3833. https://doi.org/10.3390/s22103833









