Dimensionality Reduction and Prediction of Impedance Data of Biointerface
Abstract
:1. Introduction
2. Materials and Methods
2.1. ECoG Microarrays
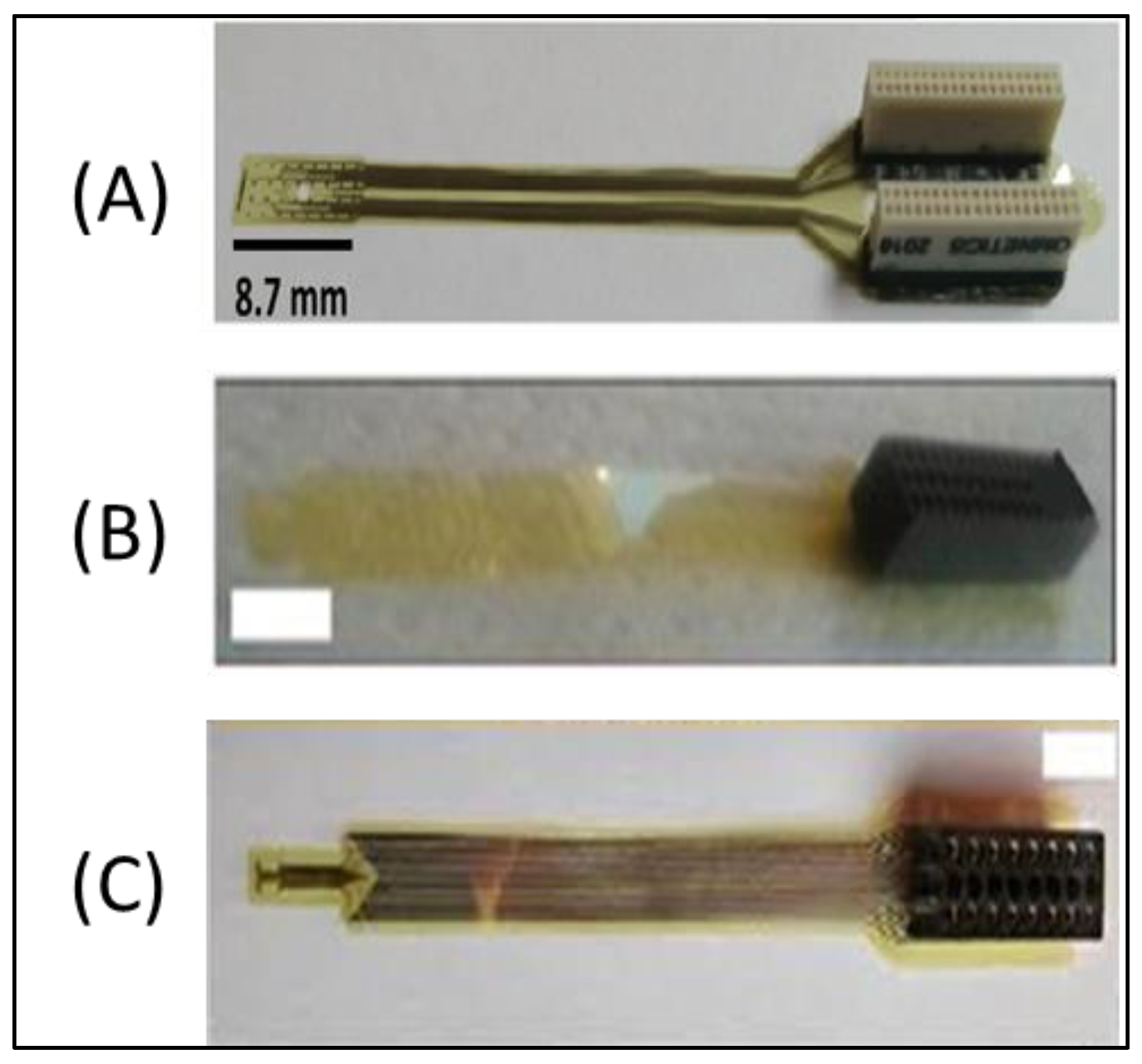
2.1.1. Polyimide-Based ECoG Microarray: “Microarray 1”
2.1.2. Polyimide-Based ECoG Microarray: “Microarray 2”
2.1.3. Polyimide-Based ECoG Microarray: “Microarray 3”
2.2. Characterization of ECoG Microarrays Using EIS
2.3. Dimensionality Reduction of Impedance Data
- : Day factor of (d) day, or normalized DF of (d) day to the first day ;
- : Day factor value of the first day;
- : Proportional values of testing frequency;
- : Impedance value at frequency;
- : Logarithm values of impedance amplitude at frequency in day (d).
2.4. Long Short-Term Memory (LSTM) Network
2.5. Performance Evaluation of Prediction
2.5.1. Accuracy of Final State (AFS)
2.5.2. Accuracy of Correlation Coefficients (ACCs)
- cov (A,B): The covariance between two random variables A and B;
- : Standard deviation of A.
3. Results
3.1. Day Factor Prediction of the Microarray 1 Impedance Data
3.2. Day factor Prediction of the Microarray 2 Impedance Data
3.3. Day Factor Prediction of the Microarray 3 Impedance Data
3.4. Comparison between the DF and PCA Approach
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lesiak-Orłowska, B. Surfaces and Interfaces in Biocatalysis. Catalysts 2022, 12, 379. [Google Scholar] [CrossRef]
- Vianello, F.; Cecconello, A.; Magro, M. Toward the Specificity of Bare Nanomaterial Surfaces for Protein Corona Formation. Int. J. Mol. Sci. 2021, 22, 7625. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Fan, Y.; Wang, M.; Cheng, Y.; Ji, B.; Chen, Y.; Wang, G. Flexible Neural Probes with Electrochemical Modified Microelectrodes for Artifact-Free Optogenetic Applications. Int. J. Mol. Sci. 2021, 22, 11528. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Lahiri, D.; Nag, M.; Dey, A.; Sarkar, T.; Pathak, S.K.; Atan Edinur, H.; Pati, S.; Ray, R.R. Bacterial Biopolymer: Its Role in Pathogenesis to Effective Biomaterials. Polymers 2021, 13, 1242. [Google Scholar] [CrossRef]
- Gayda, G.Z.; Demkiv, O.M.; Stasyuk, N.Y.; Serkiz, R.Y.; Lootsik, M.D.; Errachid, A.; Gonchar, M.V.; Nisnevitch, M. Metallic Nanoparticles Obtained via “Green” Synthesis as a Platform for Biosensor Construction. Appl. Sci. 2019, 9, 720. [Google Scholar] [CrossRef] [Green Version]
- Biru, E.I.; Necolau, M.I.; Zainea, A.; Iovu, H. Graphene Oxide–Protein-Based Scaffolds for Tissue Engineering: Recent Advances and Applications. Polymers 2022, 14, 1032. [Google Scholar] [CrossRef]
- Horváth, Á.C.; Borbély, S.; Boros, Ö.C.; Komáromi, L.; Koppa, P.; Barthó, P.; Fekete, Z. Infrared neural stimulation and inhibition using an implantable silicon photonic microdevice. Microsyst. Nanoeng. 2020, 6, 44. [Google Scholar] [CrossRef]
- Zátonyi, A.; Fedor, F.; Borhegyi, Z.; Fekete, Z. In vitro and in vivo stability of black-platinum coatings on flexible, polymer microECoG arrays. J. Neural Eng. 2018, 15, 054003. [Google Scholar] [CrossRef]
- Magar, H.S.; Hassan, R.Y.A.; Mulchandani, A. Electrochemical Impedance Spectroscopy (EIS): Principles, Construction, and Biosensing Applications. Sensors 2021, 21, 6578. [Google Scholar] [CrossRef]
- Gamal, W.; Wu, H.; Underwood, I.; Jia, J.; Smith, S.; Bagnaninchi, P.O. Impedance-based cellular assays for regenerative medicine. Phil. Trans. R. Soc. 2018, 373, 20170226. [Google Scholar] [CrossRef] [Green Version]
- Morgan, K.; Gamal, W.; Samuel, K.; Morley, S.D.; Hayes, P.C.; Bagnaninchi, P.; Plevris, J.N. Application of Impedance-Based Techniques in Hepatology Research. J. Clin. Med. 2020, 9, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morin, M.; Ruzgas, T.; Svedenhag, P.; Anderson, C.D.; Ollmar, S.; Engblom, J.; Björklund, S. Skin hydration dynamics investigated by electrical impedance techniques in vivo and in vitro. Sci. Rep. 2020, 10, 18218. [Google Scholar] [CrossRef] [PubMed]
- Fekete, Z.; Pongrácz, A. Multifunctional soft implants to monitor and control neural activity in the central and peripheral nervous system: A review. Sen. Actu. B Chem. 2017, 243, 1214–1223. [Google Scholar] [CrossRef]
- Munge, A.; Sankar, V.; Sendi, M.S.; Ghovanloo, M.; Guler, U. A bio-impedance measurement IC for neural interface applications. In Proceedings of the 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS), Cleveland, OH, USA, 17–19 October 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Iwagami, T.; Tani, T.; Ito, K.; Nishino, S.; Harashima, T.; Kino, H.; Kiyoyama, K.; Tanaka, T. Area-Efficient and Wide-Range Impedance Analysis Circuit for Multichannel High Quality Brain Signal Recording System. In Proceedings of the 2015 International Conference on Solid State Devices and Materials, Sapporo Convention Center, Sapporo, Japan, 27–30 September 2015; pp. 822–823. [Google Scholar] [CrossRef]
- Krishnan, A.; Weigle, H.; Kelly, S.; Grover, P. Feedback-based Electrode Rehydration for High Quality, Long Term, Noninvasive Biopotential Measurements and Current Delivery. In Proceedings of the 2019 IEEE Biomedical Circuits and Systems Conference (BioCAS), Nara, Japan, 17–19 October 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Lackovi, I.; Stare, Z.; Protulipac, T. On the applicability of electrical impedance indices to characterize the condition of the oral mucosa. WIT Trans. Biomed. Health 2003, 6, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Cavalieri, R.; Bertemes-Filho, P. Dimensionality reduction methods for Impedance Spectroscopy data of biological materials. In Proceedings of the 4th Latin American Conference on Bioimpedance 2021 (CLABIO 2021), San Luis Potosí, Mexico, 10–13 November 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Duan, Y.; Tian, J.; Lu, J.; Wang, C.; Shen, W.; Xiong, R. Deep neural network battery impedance spectra prediction by only using constant-current curve. Energy Storage Mater. 2021, 41, 24–31. [Google Scholar] [CrossRef]
- Komijani, H.; Rezaeihassanabadi, S.; Parsaei, M.R.; Maleki, S. Radial basis function neural network for electrochemical impedance prediction at presence of corrosion inhibitor. Peri. Polyt. Chem. Eng. 2017, 61, 128–132. [Google Scholar] [CrossRef] [Green Version]
- Caponetto, R.; Guarnera, N.; Matera, F.; Privitera, E.; Xibilia, M.G. Application of Electrochemical Impedance Spectroscopy for prediction of Fuel Cell degradation by LSTM neural networks. In Proceedings of the 2021 29th Mediterranean Conference on Control and Automation (MED), Puglia, Italy, 22–25 June 2021; pp. 1064–1069. [Google Scholar] [CrossRef]
- Csernyus, B.; Szabó, Á.; Fiáth, R.; Zátonyi, A.; Lázár, C.; Pongrácz, A.; Fekete, Z. A multimodal, implantable sensor array and measurement system to investigate the suppression of focal epileptic seizure using hypothermia. J. Neural. Eng. 2021, 18, 0460c3. [Google Scholar] [CrossRef]
- Zátonyi, A.; Borhegyi, Z.; Cserpán, D.; Somogyvári, Z.; Srivastava, M.; Kisvárday, Z.; Fekete, Z. Optical Imaging of Intrinsic Neural Signals and Simultaneous MicroECoG Recording Using Polyimide Implants. Proceedings 2017, 1, 610. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Jiang, W.; Li, Z.; Lu, Y. A New Multi-Scale Sliding Window LSTM Framework (MSSW-LSTM): A Case Study for GNSS Time-Series Prediction. Remote Sens. 2021, 13, 3328. [Google Scholar] [CrossRef]
- Cai, C.; Tao, Y.; Zhu, T.; Deng, Z. Short-Term Load Forecasting Based on Deep Learning Bidirectional LSTM Neural Network. Appl. Sci. 2021, 11, 8129. [Google Scholar] [CrossRef]
- The MathWorks, I. Deep Learning Toolbox. 2022. Available online: https://www.mathworks.com/help/deeplearning/index.html (accessed on 14 March 2022).
- Scholz, M. Approaches to Analyse and Interpret Biological Profile Data. Doctoral dissertation, Universität Potsdam, Potsdam, Germany, 2006. Available online: https://publishup.uni-potsdam.de/files/696/scholz_diss.pdf (accessed on 1 April 2022).
- Shi, Y.; Yang, Z.; Xie, F.; Ren, S.; Xu, S. The Research Progress of Electrical Impedance Tomography for Lung Monitoring. Front. Bioeng. Biotechnol. 2021, 9, 726652. [Google Scholar] [CrossRef] [PubMed]
- Cebrián-Ponce, Á.; Irurtia, A.; Carrasco-Marginet, M.; Saco-Ledo, G.; Girabent-Farrés, M.; Castizo-Olier, J. Electrical impedance myography in health and physical exercise: A systematic review and future perspectives. Front. Physiol. 2021, 12, 237496483. [Google Scholar] [CrossRef] [PubMed]
- Kolaghassi, R.; Al-Hares, M.K.; Marcelli, G.; Sirlantzis, K. Performance of Deep Learning Models in Forecasting Gait Trajectories of Children with Neurological Disorders. Sensors 2022, 22, 2969. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Park, S.; Choi, D.; Bok, K.; Yoo, J. Road Speed Prediction Scheme by Analyzing Road Environment Data. Sensors 2022, 22, 2606. [Google Scholar] [CrossRef] [PubMed]




| Reference | Dimensionality Reduction/Data/Features Extraction Tool | ML Technique for Prediction | Purpose |
|---|---|---|---|
| [17] | MIX, PIX, RIX, and IMIX | - | Diagnosing oral mucosa with a lower number of informative features. |
| [18] | PCA, LLE, mMDS, and Isomaps | MLP neural network | Dimensionality reduction of impedance data and keeping the important and informative content. |
| [19] | Constant current (CC) charging data | CNN | Predicting impedance spectra over a battery’s life. |
| [20] | Nyquist plot | RBFNN | Electrochemical impedance prediction in the presence of a corrosion inhibitor. |
| [21] | Cell voltage through cycles of FC usage | LSTM | Predicting the degradation of an FC stack. |
| ECoG Microarray | Potentiostat | Medium | RMS 1 | Soaking Days | Frequencies |
|---|---|---|---|---|---|
| Microarray 1 | nanoZ 2 | PBS 4 | 4 mV | 11 days | 1 Hz to 2 MHz |
| Microarray 2 | Gamry 3 | PBS | 25 mV | 11 days | 0.1 Hz to 10 kHz |
| Microarray 3 | Gamry | PBS | 25 mV | 11 days | 0.1 Hz to 10 kHz |
| ECoG Microarray | DF | PCA |
|---|---|---|
| Microarray 1 | 4.1 | 74.5 |
| Microarray 2 | 4.2 | 84.7 |
| Microarray 3 | 3.9 | 67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismaiel, E.; Zátonyi, A.; Fekete, Z. Dimensionality Reduction and Prediction of Impedance Data of Biointerface. Sensors 2022, 22, 4191. https://doi.org/10.3390/s22114191
Ismaiel E, Zátonyi A, Fekete Z. Dimensionality Reduction and Prediction of Impedance Data of Biointerface. Sensors. 2022; 22(11):4191. https://doi.org/10.3390/s22114191
Chicago/Turabian StyleIsmaiel, Ebrahim, Anita Zátonyi, and Zoltán Fekete. 2022. "Dimensionality Reduction and Prediction of Impedance Data of Biointerface" Sensors 22, no. 11: 4191. https://doi.org/10.3390/s22114191







