Applying Image Recognition and Tracking Methods for Fish Physiology Detection Based on a Visual Sensor
Abstract
:1. Introduction
2. Related Work
3. Problem Definition
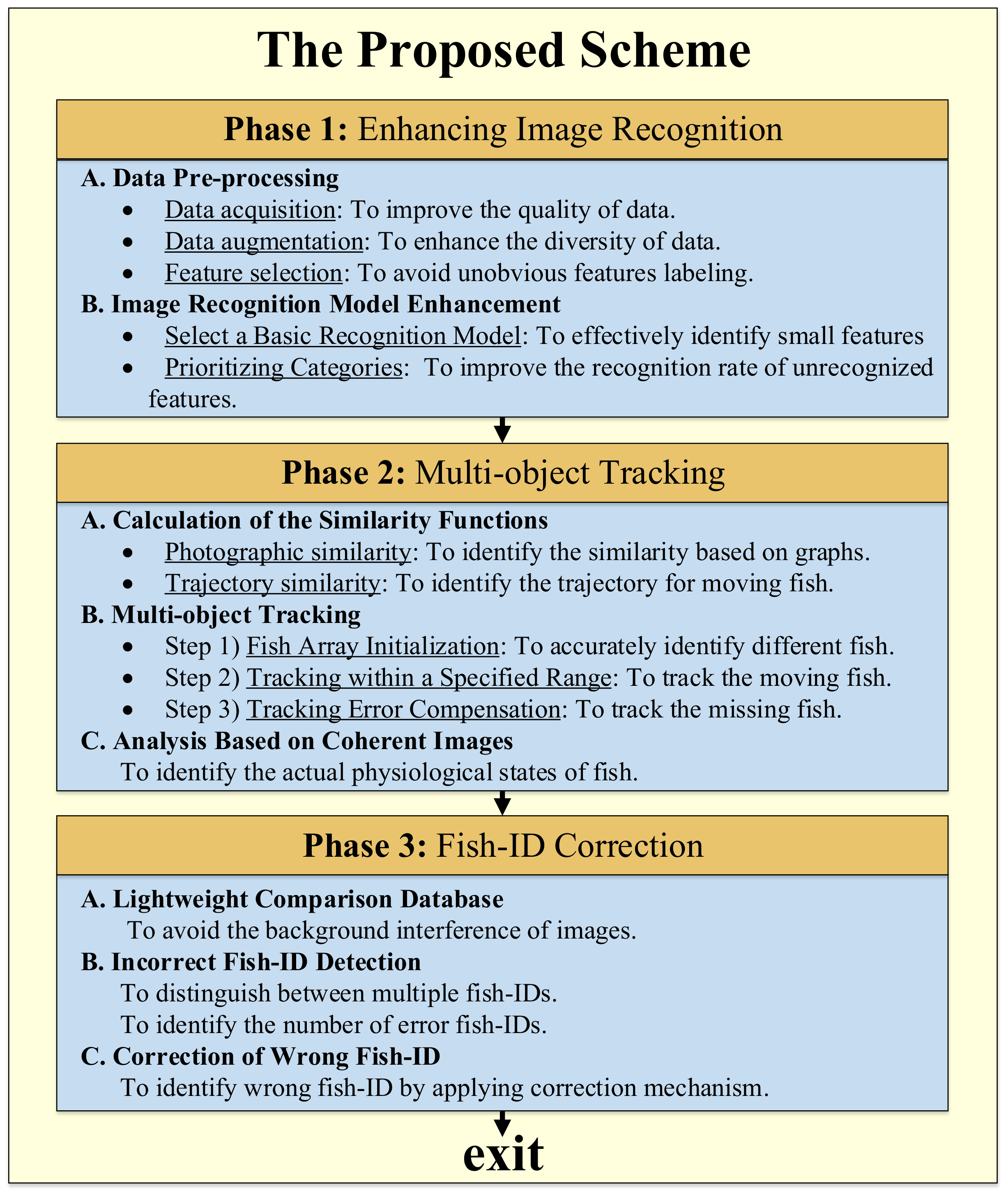
4. The Proposed Scheme
4.1. Phase 1: Enhancing Image Recognition
- A.
- Data Pre-processing
- Data acquisition: We collected more than 6000 pictures of guppies, including three physiological states: health, disease (rotten tail), and death, as shown in Figure 4. These pictures are obtained from (1) online guppy photos and (2) the real guppy photographed by the visual sensors (cameras). Note that the online guppy photos are from Taiwan Fish Database [20], Guppy Dataset [21], and other searching engine results; the real guppy pictures are photographed by the visual sensors with a pixel resolution of 4032 × 3024 and 5184 × 3880, respectively. The characteristics of the guppy pictures include their various appearances, such as front view, back view, side view, swimming up, and swimming down, under the environments of clear background and background with landscaping.
- Data augmentation: To improve the overall data diversity, we randomly select photos to process with discoloration, rotation, or mirroring to increase the amount of training data, which can make the recognition model more general and avoid overfitting [8,22,23]. As the images of fish with certain features may not be easy to obtain (such as rotten tails), we also dye the existing rotten-tail images with random colors, such as blue, green, purple, red, and black colors, which are the common colors of guppy tails, in order to balance the data of the three physiological states, as shown in Figure 5. This can potentially improve the recognition capability.
- Feature selection: According to the literature [24,25], the caudal fin of rotten-tail fish will have irregular notches or a jagged appearance. Thus, we label these features according to the above description and consult the experts working in the aquarium for guidance [26]. Sometimes, it is difficult to judge because of the irregular swimming of the fish and the angle of view, as shown in Figure 6. Therefore, we only can label the feature of the images that meet the above description, so as to avoid labeling with inconspicuous characteristics of rotten tails. Note that if the images of fish with inconspicuous features are used as training data, the tails of healthy fish are easily misidentified as rotten-tail fish, potentially resulting in a decrease in the recognition rate.
- B.
- Enhancing the Image Recognition Model
- The two-stage detection method is adopted to separate the object position detection and the object identification, so that it has a better performance in terms of accuracy. Compared with the one-stage detection method, the detection of small targets will be more accurate.
- It is more flexible and does not have too many restrictions on image input. Images with different aspect ratios are allowed, and images with different aspect ratios can also be used for training.
- The RPN (region proposal network) is exploited as the extraction network of the candidate frame, which improves the speed while improving the accuracy.
- Step (1)
- First, it extracts the feature map of the image with the pre-trained network (e.g., VGG16, ResNet), and the feature map will be shared by the subsequent RPN layer and the fully connected layer. Note that we use VGG16 as our pre-trained network, which has a better performance in small feature recognition.
- Step (2)
- Then, the RPN layer generates candidate boxes (region of interest, RoI). This layer replaces the selective search used by the previous versions of R-CNN and faster R-CNN to extract candidate boxes, and leverages the concept of anchor boxes by the operation of sliding window on the feature map. The position of the center point of the current sliding window corresponds to the pixel space of the original image, and then k anchor boxes with different sizes and different aspect ratios are generated on the original image. The size of each anchor box will vary according to the size of the input image, as shown in Figure 8. Finally, it estimates whether the anchor boxes belong to the target (positive) or not (negative) through the softmax function, and then uses the bounding box regression to adjust the position information of the anchor boxes to obtain the accurate position of candidate frames.
- Step (3)
- After that, RoI pooling is performed on the previously generated feature map and the candidate frame generated by the RPN layer. As the subsequent classification layer requires an input of the same size, RoI pooling maps candidate boxes of different sizes to the feature map and makes them the same size, and then passes the features of the same size to the subsequent classification layer for classification.
- Step (4)
- Finally, the classification layer calculates the probability that the object in each candidate frame belongs to each category through the fully connected layer and softmax, and then determines to which category the object belongs, and uses bounding box regression again to obtain a more accurate target detection frame.
- Prioritizing Categories
- To improve the recognition rate of unrecognized features, we set the recognition priority for each category and adjust the confidence thresholds of different categories.
- The identification priority is also set by the following order: dead > diseased (rotten-tail) > healthy. As the diseased (rotten-tail) fish would be recognized as healthy from certain angles, as illustrated in Figure 9, the higher priority should be given for the diseased (rotten-tail) category to avoid recognizing them as healthy. Note that the characteristics of fish death are the most obvious, so the priority is the highest.
- Next, let the threshold of the health category be , the threshold of the diseased (rotten-tail) category be , and the threshold of the death category be . Then, according to the above priority, when the confidence value of the category is greater than the corresponding threshold (e.g., , , ), it will be determined as the corresponding category.
4.2. Phase 2: Multi-Object Tracking
- A.
- Calculation of the Similarity Functions
- Photographic similarity
- Trajectory similarity
- B.
- Multi-object Tracking Design
- Step (1)
- Fish Array Initialization: First, we create a fish array A = [IDi, si, (xi, yi, wi, hi)], i = 1..N to store their IDs (initially a random number), state, and bounding box coordinate of each fish, which is identified by faster R-CNN in Phase 1.
- Step (2)
- Tracking within a Specified Range: Next, we take the center coordinate of the bounding box at the previous time (t − 1), select a specific range at the radius , and search for the center coordinates of all the bounding boxes at the current time (t) within this range. Then, we compare the bounding box of the previous moment (t − 1) with all of the identification frames of the current time (t) within the circle selection range and calculate their similarity by . After the comparison, the bounding box with the highest similarity at the previous time (t − 1) is selected, which will inherit the ID of the former time, and replace it with the initial ID, and then update the tracking array accordingly.
- Step (3)
- Tracking Error Compensation: In the following tracking process, if there are fish that fail to be detected, or the bounding box is missing because of shading with each other, we design a tracking error compensation mechanism to record the relevant information at the moment (t − 1) before the ID is missing. Once the ID tracking is performed at the next moment (t + 1), the range of the search radius is expanded by Ω times for ID exploration, so as to avoid the tracking failure caused by the fish moving far away. Note that when the system is started, the number of fish will be matched to the number of bounding boxes. If the fish disappears over frames, the current missing ID will be stored. Once the fish appears again, the missing ID of the fish will be assigned.
- C.
- Analysis Based on Coherent Images
4.3. Phase 3: Fish-ID Correction
- A.
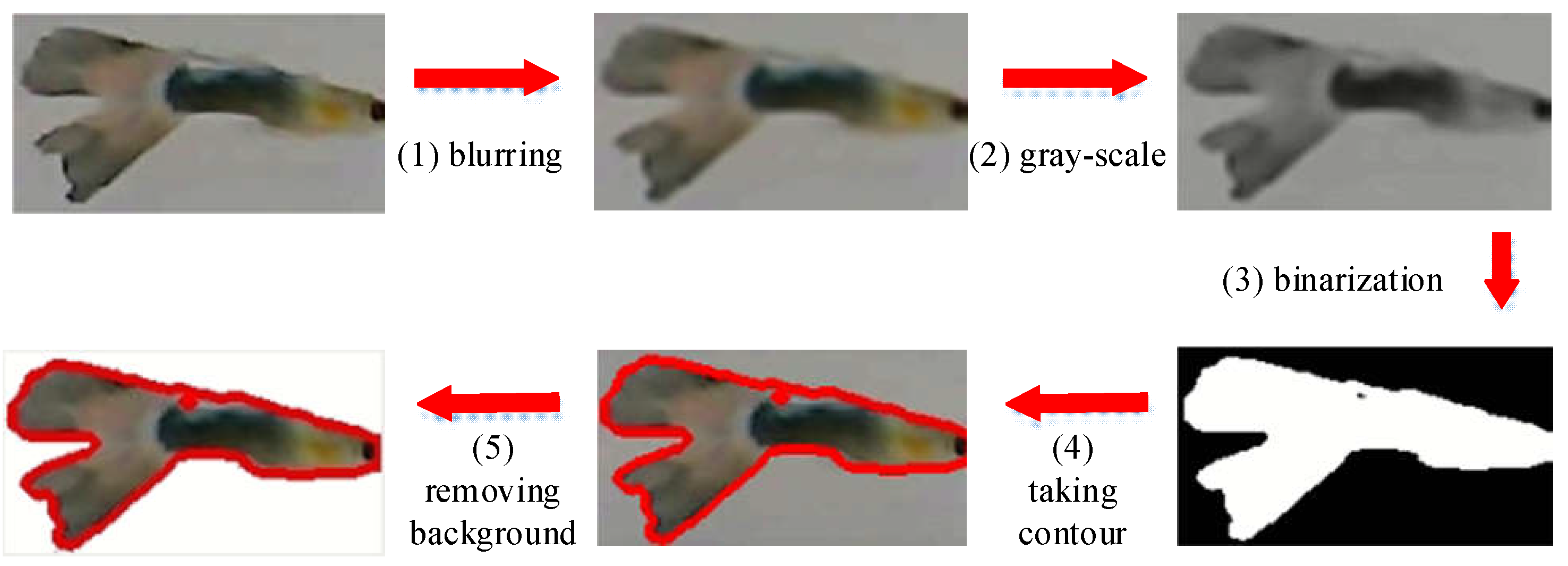
- Establishment of a Lightweight Comparison Database: According to the ID within frames after the system starts, each fish is intercepted in λ valid images and added to the fish comparison database. Then, we expand these images through mirroring and flipping [8]. After that, we have 2λ images for each fish-ID i, denoted by Q(i). Here, to avoid the background interference of fish images, we perform the following image processing processes [32], including (1) blurring, (2) gray-scale, (3) binarization (black and white), (4) taking contours, (5) removing background, and then storing the images in Q(i), as illustrated in Figure 12. In this way, the interference caused by the background can be effectively reduced.
- B.
- Incorrect Fish-ID Detection
- Step (1)
- For the fish images stored in Q(i), the BGR histogram of R(i) is generated by the OpenCV function (calcHist).
- Step (2)
- Then, we use the function (compareHist) to perform a histogram coincidence comparison between the image in the database Q(i) and the target image R(i).
- Step (3)
- Next, we compare the target image R(i) with the λ images in Q(i) and calculate the average value based on the histogram gravity and degree H(i). If the calculated result exceeds the pre-defined threshold (), it means that the image similarity of the target ID is low.
- Step (4)
- To avoid a false alarm caused by temporary low similarity such as fish turning around or shading with each other, it will observe the similarity through frames. Once the consecutive frames all have low similarity, it is identified as a wrong ID.
- C.
- Correction of Wrong Fish-ID
- The above incorrect fish-ID detection will be activated periodically by frames. Once it detects at least two wrong IDs, it will activate the correction mechanism.
- Assuming that the total number of wrong IDs is Φ ≥ 2, the correction mechanism will cross-match all the wrong IDs’ images in the comparison database Q(i).
- If the IDs of the two most similar images are matched, the wrong IDs will be updated by the ID in the comparison database. The operation will be executed continuously until the number of wrong IDs is less than 2.
5. Performance Evaluation
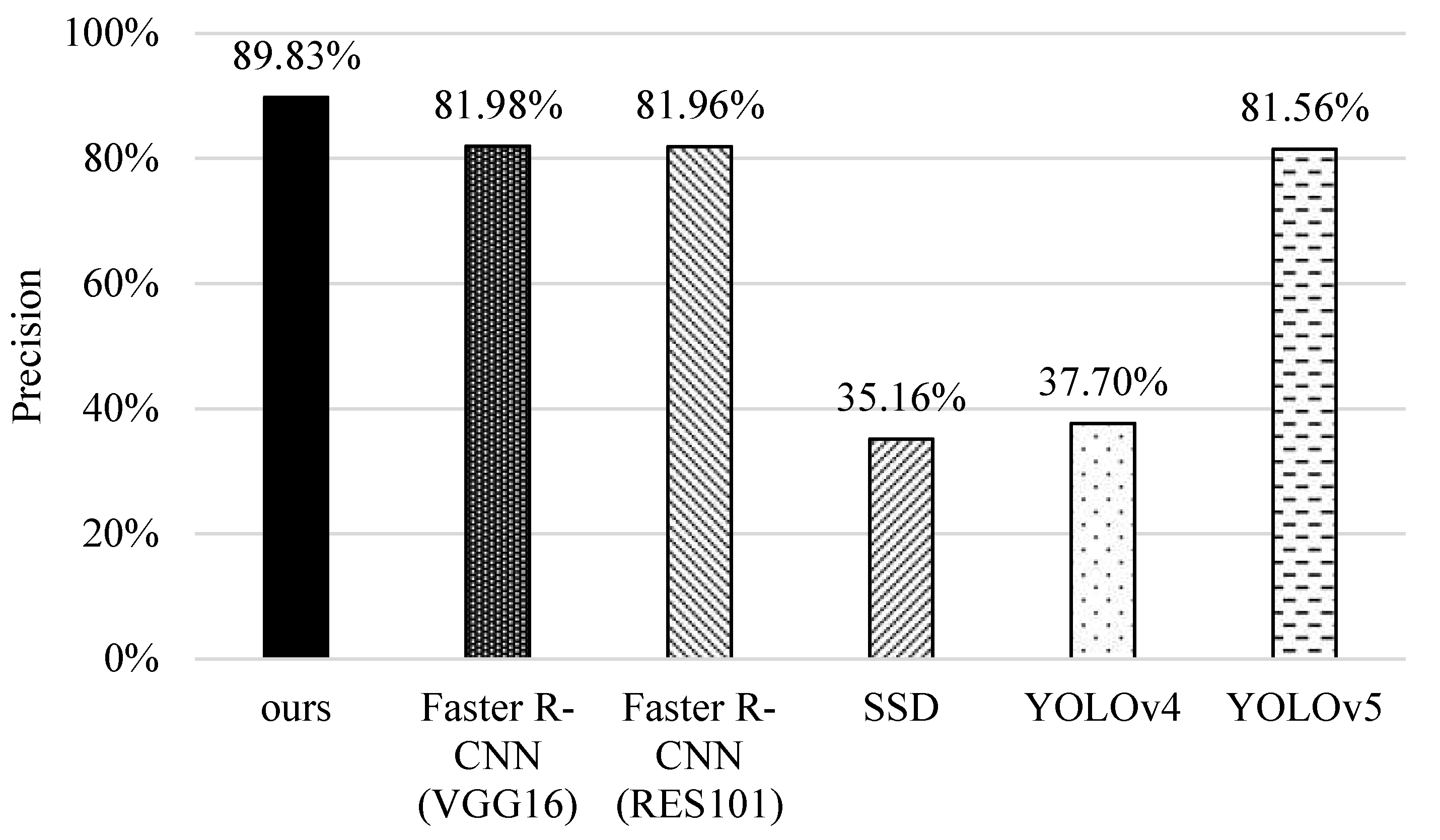
5.1. Precision
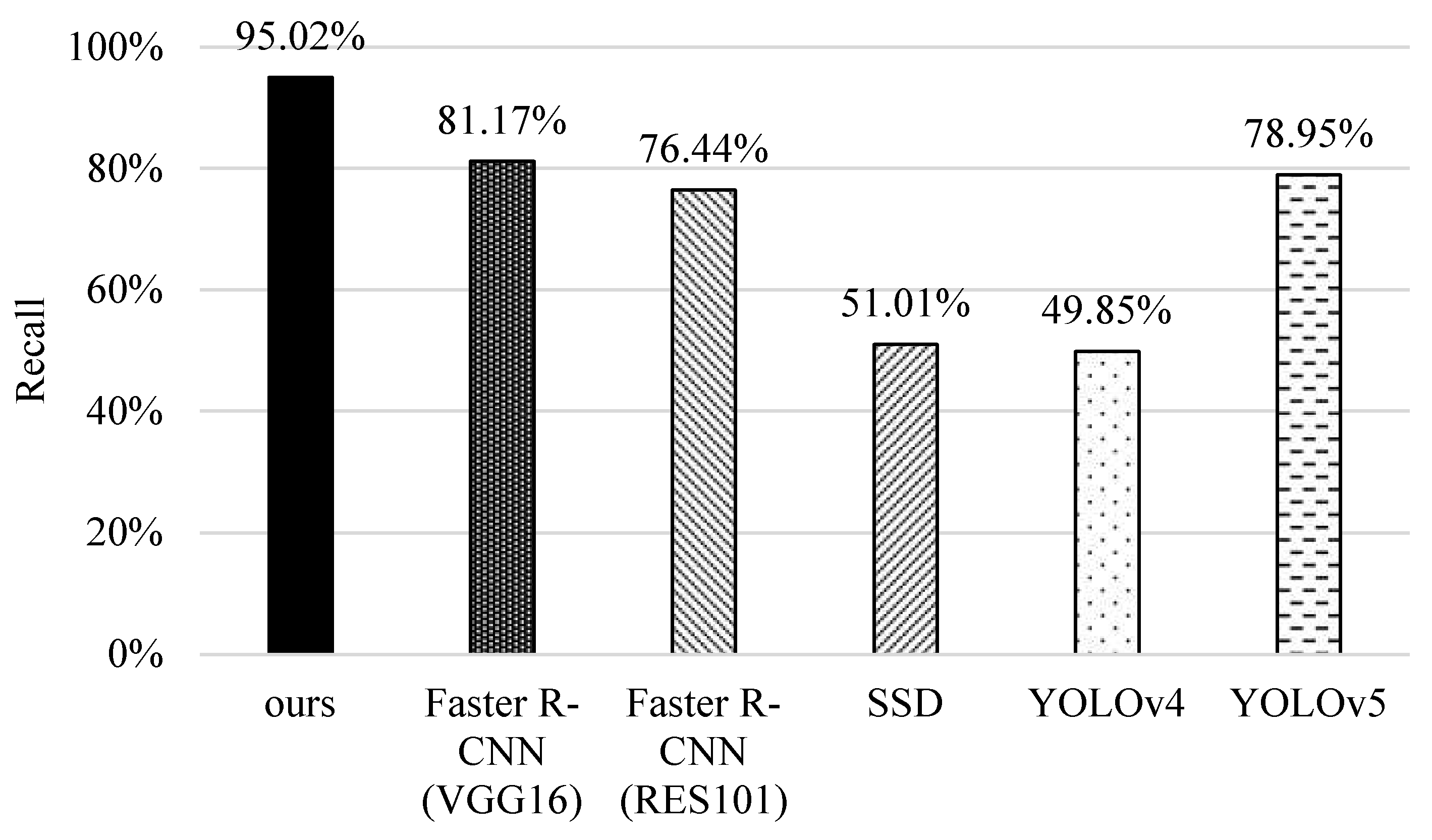
5.2. Recall
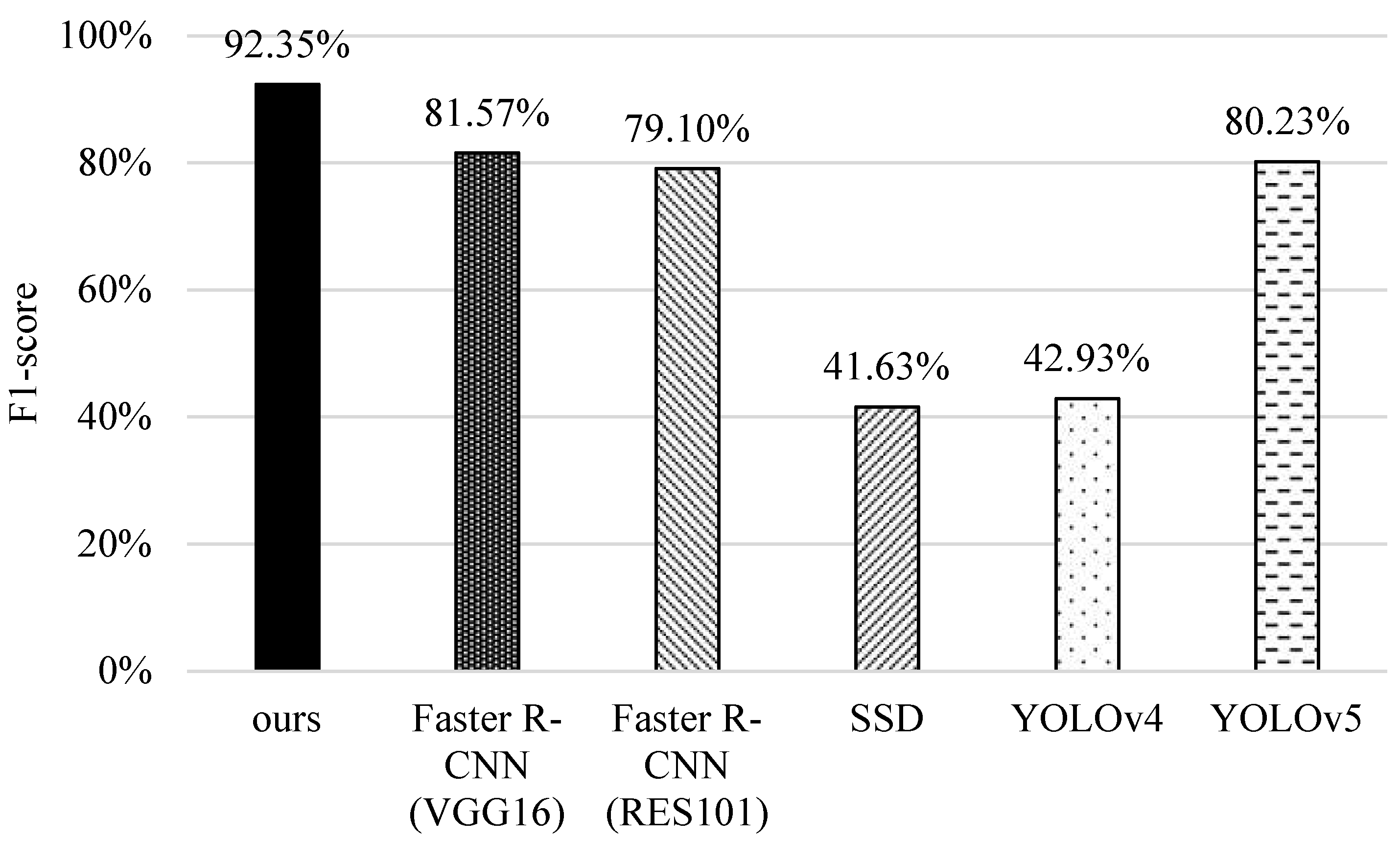
5.3. F1-Score
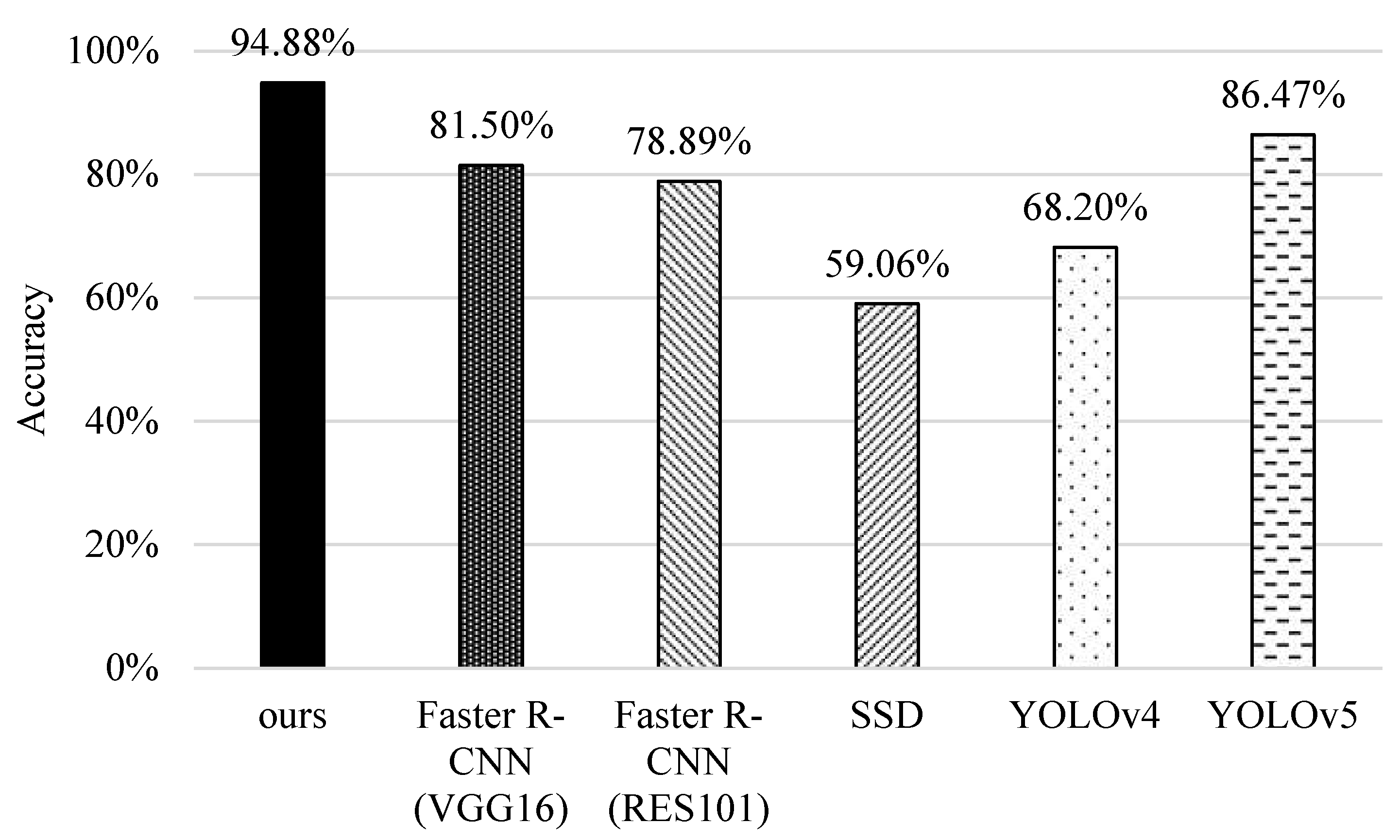
5.4. Accuracy
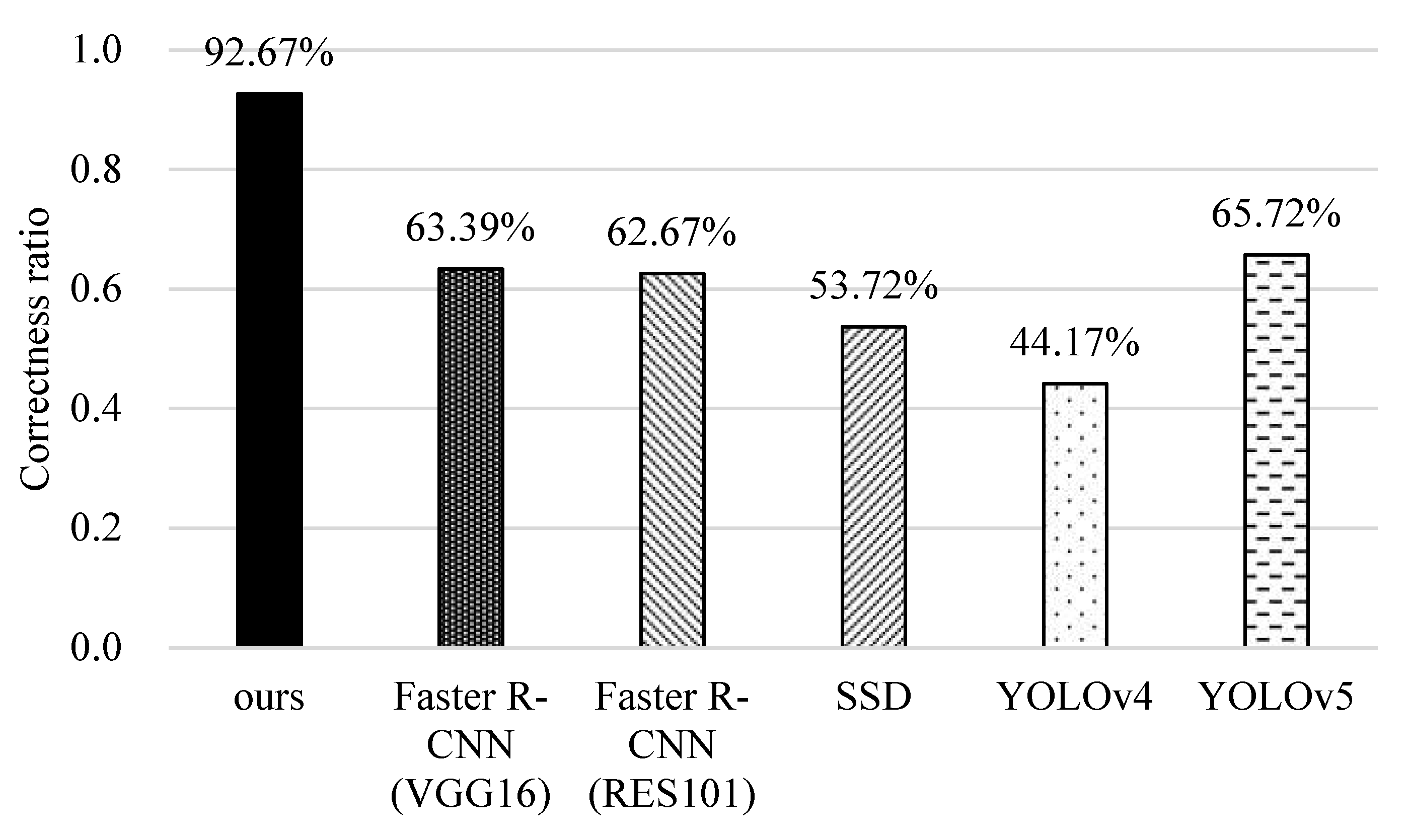
5.5. Identification of Unknown Data Lists
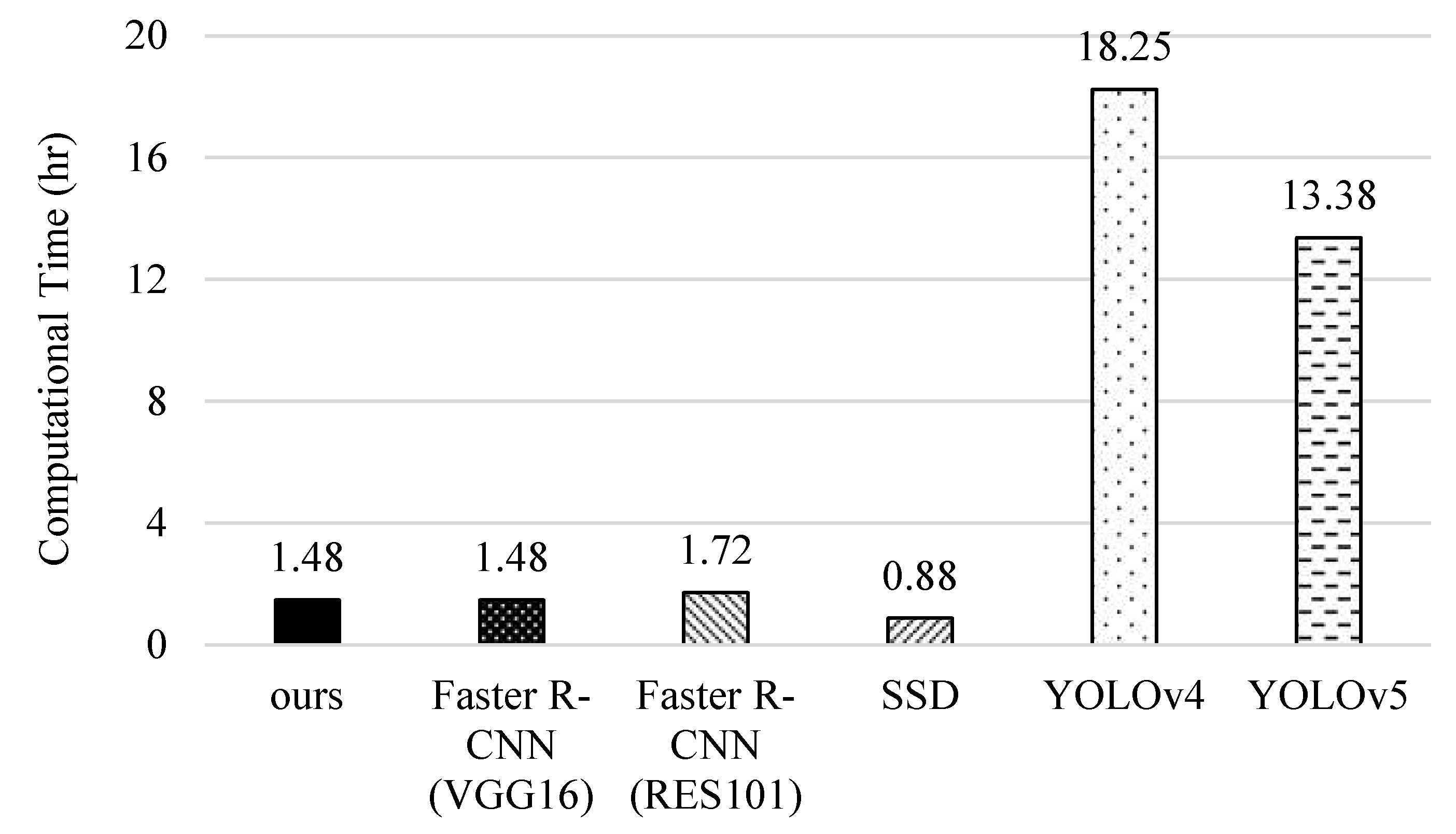
5.6. Computational Complexity
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Business Next. Available online: https://www.bnext.com.tw/article/59088/pet-market-2020 (accessed on 2 September 2020).
- Fish Keeping World. Available online: https://www.fishkeepingworld.com/15-ways-fish-reduce-stress-and-improve-mental-health/ (accessed on 15 July 2022).
- Nimer, J.; Lundahl, B. Animal-Assisted Therapy: A Meta-Analysis. Anthrozoös 2015, 20, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Rajib, R.H.; Kabir, S. IOT Based Automated Fish Feeder. In Proceedings of the International Conference on Innovations in Science, Engineering and Technology (ICISET), Pune, India, 13–15 February 2020; pp. 90–93. [Google Scholar]
- Daud, A.K.P.M.; Sulaiman, N.A.; Yusof, Y.W.M.; Kassim, M. An IoT-Based Smart Aquarium Monitoring System. In Proceedings of the IEEE Symposium on Computer Applications & Industrial Electronics (ISCAIE), Malaysia, 18–19 April 2020; pp. 277–282. [Google Scholar]
- Sharmin, I.; Islam, N.F.; Jahan, I.; Joye, T.A.; Rahman, R.; Habib, T. Machine Vision Based Local Fish Recognition. Appl. Sci. 2019, 1, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Park, J.H.; Choi, Y.K. Efficient Data Acquisition and CNN Design for Fish Species Classification in Inland Waters. Inf. Commun. Converg. Eng. 2020, 18, 106–114. [Google Scholar]
- Montalbo, F.J.P.; Hernandez, A.A. Classification of Fish Species with Augmented Data Using Deep Convolutional Neural Network. In Proceedings of the IEEE International Conference on System Engineering and Technology (ICSET), Shah Alam, Malaysia, 7 October 2019; pp. 396–401. [Google Scholar]
- Sung, M.; Yu, S.C.; Girdhar, Y. Vision Based Real-Time Fish Detection Using Convolutional Neural Network. In Proceedings of the IEEE OCEANS, Anchorage, AK, USA, 18–21 September 2017; pp. 1–6. [Google Scholar]
- Wang, H.; Shi, Y.; Yue, Y.; Zhao, H. Study on Freshwater Fish Image Recognition Integrating SPP and DenseNet Network. In Proceedings of the IEEE International Conference on Mechatronics and Automation (ICMA), Beijing, China, 13–16 October 2020; pp. 1–6. [Google Scholar]
- Rauf, H.T.; Lali, M.I.U.; Zahoor, S.; Shah, S.Z.H.; Rehman, A.U.; Bukhari, S.A.C. Visual Features Based Automated Identification of Fish Species Using Deep Convolutional Neural Networks. Comput. Electron. Agric. 2019, 167, 1–17. [Google Scholar] [CrossRef]
- Huang, Y.; Jin, S.; Fu, S.; Meng, C.; Jiang, Z.; Ye, S.; Yang, D.; Li, R. Aeromonas Hydrophila as A Causative Agent of Fester-Needle Tail Disease in Guppies (Poecilia Reticulata). Int. J. Agric. Biol. 2021, 15, 397–403. [Google Scholar] [CrossRef]
- Hossain, S.; de Silva, B.C.J.; Dahanayake, P.S.; de Zoysa, M.; Heo, G.J. Phylogenetic Characteristics, Virulence Properties and Antibiogram Profile of Motile Aeromonas Spp. Isolated from Ornamental Guppy (Poecilia Reticulata). Arch. Microbiol. 2020, 202, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Guo, C.; Zheng, X.; Yu, Z.; Wang, W.; Zheng, H.; Fu, M.; Zheng, B. Fish Recognition from a Vessel Camera Using Deep Convolutional Neural Network and Data Augmentation. In Proceedings of the IEEE Kobe Techno-Oceans (OTO), Kobe, Japan, 28–31 May 2018; pp. 1–5. [Google Scholar]
- Chen, G.; Sun, P.; Shang, Y. Automatic Fish Classification System Using Deep Learning. In Proceedings of the IEEE International Conference on Tools with Artificial Intelligence (ICTAI), Boston, MA, USA, 6–8 November 2017; pp. 24–29. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2016; pp. 21–37. [Google Scholar]
- Santos, A.A.d.; Gonçalves, W.N. Improving Pantanal Fish Species Recognition through Taxonomic Ranks in Convolutional Neural Networks. Ecol. Inform. 2019, 53, 1–11. [Google Scholar] [CrossRef]
- Siddiqui, S.A.; Salman, A.; Malik, M.I.; Shafait, F.; Mian, A.; Shortis, M.R.; Harvey, E.S. Automatic Fish Species Classification in Underwater Videos: Exploiting Pre-Trained Deep Neural Network Models to Compensate for Limited Labelled Data. J. Mar. Sci. 2018, 75, 374–389. [Google Scholar] [CrossRef]
- Saengsitthisak, B.; Chaisri, W.; Punyapornwithaya, V.; Mektrirat, R.; Bernard, J.K.; Pikulkaew, S. The Current state of Biosecurity and Welfare of Ornamental Fish Population in Pet Fish Stores in Chiang Mai Province, Thailand. Vet. Integr. Sci. 2021, 19, 277–294. [Google Scholar] [CrossRef]
- Taiwan Fish Database. Available online: https://fishdb.sinica.edu.tw/chi/species.php?id=381035; https://fishdb.sinica.edu.tw/eng/specimenlist3.php?m=ASIZ&D1=equal&R1=science&T3=Poecilia%20reticulata (accessed on 15 July 2022).
- Guppy Dataset (Male guppy (Poecilia reticulata) photographs from five sites in the Guanapo tributary in Trinidad). Available online: https://figshare.com/articles/dataset/Male_guppy_Poecilia_reticulata_photographs_from_five_sites_in_the_Guanapo_tributary_in_Trinidad_/6130352 (accessed on 15 July 2022).
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Kaur, P.; Khehra, B.S.; Mavi, E.B.S. Data Augmentation for Object Detection: A Review. In Proceedings of the IEEE International Midwest Symposium on Circuits and Systems (MWSCAS), East Lansing, MI, USA, 8–11 August 2021; pp. 537–543. [Google Scholar]
- Roberts, H.E.; Palmeiro, B.; Weber, E.S. Bacterial and Parasitic Diseases of Pet Fish. Vet. Clin. N. Am. Exot. Anim. Pract. 2009, 12, 609–638. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, P.H.M.; Moreno, A.M.; Moreno, L.Z.; Oliveira, C.H.; Baroni, F.A.; Maganha, S.R.L.; Sousa, R.L.M.; Balian, S.C. Infectious Diseases in Aquarium Ornamental Pet Fish: Prevention and Control Measures. Braz. J. Vet. Res. Anim. Sci. 2019, 56, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Rainbow Fish Pet Aquarium. Available online: https://032615588.web66.com.tw/ (accessed on 15 July 2022).
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 137–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, K.; Yang, H.-F.; Hsiao, J.-H.; Chen, C.-S. Deep Learning of Binary Hash Codes for Fast Image Retrieval. In Proceedings of the Computer Vision and Pattern Recognition Workshops (CVPRW), Boston, MA, USA, 7–12 June 2015; pp. 27–35. [Google Scholar]
- Cao, W.; Feng, W.; Lin, Q.; Cao, G.; He, Z. A Review of Hashing Methods for Multimodal Retrieval. IEEE Access 2020, 8, 15377–15391. [Google Scholar] [CrossRef]
- Masek, W.; Paterson, M.A. A Faster Algorithm Computing String Edit Distances. J. Comput. Syst. Sci. 1980, 20, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, B. A Normalized Levenshtein Distance Metric. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 1091–1095. [Google Scholar]
- Yan, X.; Jing, G.; Cao, M.; Zhang, C.; Liu, Y.; Wang, X. Research of Sub-pixel Inner Diameter Measurement of Workpiece Based on OpenCV. In Proceedings of the 2018 International Conference on Robots & Intelligent System (ICRIS), Amsterdam, Netherlands, 21–23 February 2018; pp. 1–4. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Jocher, G.; Nishimura, K.; Mineeva, T.; Vilariño, R. YOLOv5. Available online: https://github.com/ultralytics/yolov5 (accessed on 10 July 2020).
- Chao, W.-L. Machine Learning Tutorial. Digital Image and Signal Processing; National Taiwan University: Taiwan, 2011. [Google Scholar]
- Raschka, S.; Mirjalili, V. Python Machine Learning: Machine Learning and Deep Learning with Python, Scikit-learn, and TensorFlow 2; Packt Publishing Ltd: Birmingham, UK, 2019. [Google Scholar]










| Labeled Data List<Bf, If, Sf> | |
| B1 | ((x1, y1, l1, w1), (x2, y2, l2, w2), (x3, y3, l3, w3), (x4, y4, l4, w4), (x5, y5, l5, w5), …) |
| I1 | (Nemo, Bob, Mary, Alice, Tom…) |
| S1 | (H, R, R, D, H, …) |
| B2 | ((x1, y1, l1, w1), (x2, y2, l2, w2), (x3, y3, l3, w3), (x4, y4, l4, w4), (x5, y5, l5, w5), …) |
| I2 | (Bob, Mary, Alice, Tom, Nemo…) |
| S2 | (R, R, D, H, H …) |
| B3 | ((x1, y1, l1, w1), (x2, y2, l2, w2), (x3, y3, l3, w3), (x4, y4, l4, w4), (x5, y5, l5, w5), …) |
| I3 | (Mary, Nemo, Bob, Alice, Tom…) |
| S3 | (R, H, R, D, H…) |
| B4 | ((x1, y1, l1, w1), (x2, y2, l2, w2), (x3, y3, l3, w3), (x4, y4, l4, w4), (x5, y5, l5, w5), …) |
| I4 | (Bob, Mary, Alice, Tom, Nemo…) |
| S4 | (R, R, D, H, H…) |
| B5 | ((x1, y1, l1, w1), (x2, y2, l2, w2), (x3, y3, l3, w3), (x4, y4, l4, w4), (x5, y5, l5, w5), …) |
| I5 | (Tom, Nemo, Bob, Mary, Alice…) |
| S5 | (H, H, R, R, D…) |
| … | … |
| Unknown Data List(Bx, Ix, Sx) | |
| Bx | ((?, ?, ?, ?), (x2, y2, w2, h2), (?,?,?,?), (x4, y4, w4, h4), (?,?,?,?), …) |
| Ix | (Bob, ?, Alice, ?, …, …) |
| Sx | (?, R, ?, D, ?, …) |
| Parameter | Ours | Faster-RCNNs | SSD | YOLOv4/v5 |
|---|---|---|---|---|
| learning rate | 0.001~0.0001 | 0.001~0.0001 | 0.001~0.0001 | 0.01~0.001 |
| batch size | 256 | 256 | 8 | 64/16 |
| momentum | 0.9 | 0.9 | 0.9 | 0.949/0.98 |
| weight decay | 0.0005 | 0.0005 | 0.0005 | 0.0005/0.001 |
| Parameters of the proposed scheme | ||||
| ) = (0.5, 0.3, 0.9) | = 5 | |||
| = image length/3 | = 9 | |||
| Ω = 1.5 | = 20 | |||
| = 5 | = 5 | |||
| = 4 | = 0.14 | |||
| = 10 | α = 0.15 | |||
| Actual Class\Predicted Class | Positive | Negative |
|---|---|---|
| Positive | TP | FN |
| Negative | FP | TN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, J.-M.; Mishra, S.; Cheng, Y.-L. Applying Image Recognition and Tracking Methods for Fish Physiology Detection Based on a Visual Sensor. Sensors 2022, 22, 5545. https://doi.org/10.3390/s22155545
Liang J-M, Mishra S, Cheng Y-L. Applying Image Recognition and Tracking Methods for Fish Physiology Detection Based on a Visual Sensor. Sensors. 2022; 22(15):5545. https://doi.org/10.3390/s22155545
Chicago/Turabian StyleLiang, Jia-Ming, Shashank Mishra, and Yu-Lin Cheng. 2022. "Applying Image Recognition and Tracking Methods for Fish Physiology Detection Based on a Visual Sensor" Sensors 22, no. 15: 5545. https://doi.org/10.3390/s22155545
APA StyleLiang, J. -M., Mishra, S., & Cheng, Y. -L. (2022). Applying Image Recognition and Tracking Methods for Fish Physiology Detection Based on a Visual Sensor. Sensors, 22(15), 5545. https://doi.org/10.3390/s22155545







