A Plasmid-Based Fluorescence Reporter System for Monitoring Oxidative Damage in E. coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Strains and Standard Cultivation Conditions
2.2. Assembly of the Reporter Plasmids and the Corresponding Expression Strains
2.3. Oxidative Stress Analysis of the Reporter Strains Using a Microplate Reader
2.4. Sample Preparation for CellROX Comparison
2.5. Image Acquisition Using Confocal Fluorescence Microscopy for ROS Analysis
2.6. Image Analysis and Quantification of ROS-Mediated Effects
3. Results
3.1. Design of a Fluorescent Sensor System for Measuring Oxidative Stress Based on IscR
3.2. Dynamic Fluorescence Stress Response Induced by Supplemented H2O2
3.3. Lowered Cultivation Temperature Triggers an IscR-Mediated Fluorescent Stress Response
3.4. Comparison of the Reporter System with a Commercial Probe Sensitive towards Oxidising Agents
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| E. coli | Escherichia coli |
| Em | Emission |
| Ex | Excitation |
| IPTG | Isopropyl β-d-1-thiogalactopyranoside |
| IscR | Iron–sulphur cluster biosynthesis regulator protein |
| iscR | The gene encoding the IscR protein |
| LSM | Laser scanning microscope |
| OD | Optical density |
| ROS | Reactive oxygen species |
| rpm | Revolutions per minute |
References
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, B. Biochemistry of oxidative stress. Biochem. Soc. Trans. 2007, 35, 1147–1150. [Google Scholar] [CrossRef] [PubMed]
- Khorobrykh, S.; Havurinne, V.; Mattila, H.; Tyystjärvi, E. Oxygen and ROS in Photosynthesis. Plants 2020, 9, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imlay, J.A. How oxygen damages microbes: Oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 2002, 46, 111–153. [Google Scholar]
- Khademian, M.; Imlay, J.A. How microbes evolved to tolerate oxygen. Trends Microbiol. 2021, 29, 428–440. [Google Scholar] [CrossRef]
- Schwartz, C.J.; Giel, J.L.; Patschkowski, T.; Luther, C.; Ruzicka, F.J.; Beinert, H.; Kiley, P.J. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 14895–14900. [Google Scholar] [CrossRef] [Green Version]
- Imlay, J.A. Transcription factors that defend bacteria against reactive oxygen species. Annu. Rev. Microbiol. 2015, 69, 93–108. [Google Scholar] [CrossRef] [Green Version]
- Mettert, E.L.; Kiley, P.J. Coordinate regulation of the Suf and Isc Fe-S cluster biogenesis pathways by IscR is essential for viability of Escherichia coli. J. Bacteriol. 2014, 196, 4315–4323. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.A.; Pereira, P.J.B.; Macedo-Ribeiro, S. What a difference a cluster makes: The multifaceted roles of IscR in gene regulation and DNA recognition. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2015, 1854, 1101–1112. [Google Scholar] [CrossRef]
- Blanc, B.; Gerez, C.; de Choudens, S.O. Assembly of Fe/S proteins in bacterial systems: Biochemistry of the bacterial ISC system. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2015, 1853, 1436–1447. [Google Scholar] [CrossRef]
- Giel, J.L.; Rodionov, D.; Liu, M.; Blattner, F.R.; Kiley, P.J. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 2006, 60, 1058–1075. [Google Scholar] [CrossRef]
- Jang, S.; Imlay, J.A. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol. Microbiol. 2010, 78, 1448–1467. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Shin, J.H.; Lee, K.L.; Imlay, J.A.; Roe, J.H. Comparative study of SoxR activation by redox-active compounds. Mol. Microbiol. 2013, 90, 983–996. [Google Scholar] [CrossRef] [Green Version]
- Wardman, P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: Progress, pitfalls, and prospects. Free Radic Biol. Med. 2007, 43, 995–1022. [Google Scholar] [CrossRef]
- Imlay, J.A. Diagnosing oxidative stress in bacteria: Not as easy as you might think. Curr. Opin. Microbiol. 2015, 24, 124–131. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Tian, X.; Shin, I.; Yoon, J. Fluorescent and luminescent probes for detection of reactive oxygen and nitrogen species. Chem. Soc. Rev. 2011, 40, 4783–4804. [Google Scholar] [CrossRef]
- Gomes, A.; Fernandes, E.; Lima, J.L. Fluorescence probes used for detection of reactive oxygen species. J. Biochem. Biophys. Methods 2005, 65, 45–80. [Google Scholar] [CrossRef]
- Bardelang, D.; Rockenbauer, A.; Karoui, H.; Finet, J.P.; Biskupska, I.; Banaszak, K.; Tordo, P. Inclusion complexes of EMPO derivatives with 2,6-di-O-methyl-beta-cyclodextrin: Synthesis, NMR and EPR investigations for enhanced superoxide detection. Org. Biomol. Chem. 2006, 4, 2874–2882. [Google Scholar] [CrossRef]
- Kalyanaraman, B.; Darley-Usmar, V.; Davies, K.J.; Dennery, P.A.; Forman, H.J.; Grisham, M.B.; Mann, G.E.; Moore, K.; Roberts, L.J.; Ischiropoulos, H. Measuring reactive oxygen and nitrogen species with fluorescent probes: Challenges and limitations. Free Radic Biol. Med. 2012, 52, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Afzal, M.; Matsugo, S.; Sasai, M.; Xu, B.; Aoyama, K.; Takeuchi, T. Method to overcome photoreaction, a serious drawback to the use of dichlorofluorescin in evaluation of reactive oxygen species. Biochem. Biophys. Res. Commun. 2003, 304, 619–624. [Google Scholar] [CrossRef]
- Setsukinai, K.; Urano, Y.; Kakinuma, K.; Majima, H.J.; Nagano, T. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J. Biol. Chem. 2003, 278, 3170–3175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchesi, E.; Rota, C.; Fann, Y.C.; Chignell, C.F.; Mason, R.P. Photoreduction of the fluorescent dye 2′-7′-dichlorofluorescein: A spin trapping and direct electron spin resonance study with implications for oxidative stress measurements. Free Radic. Biol. Med. 1999, 26, 148–161. [Google Scholar] [CrossRef]
- Wrona, M.; Patel, K.; Wardman, P. Reactivity of 2′, 7′-dichlorodihydrofluorescein and dihydrorhodamine 123 and their oxidized forms toward carbonate, nitrogen dioxide, and hydroxyl radicals. Free Radic. Biol. Med. 2005, 38, 262–270. [Google Scholar] [CrossRef]
- Winterbourn, C.C. The challenges of using fluorescent probes to detect and quantify specific reactive oxygen species in living cells. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2014, 1840, 730–738. [Google Scholar] [CrossRef]
- Cheng, G.; Zielonka, M.; Dranka, B.; Kumar, S.N.; Myers, C.R.; Bennett, B.; Garces, A.M.; Dias Duarte Machado, L.G.; Thiebaut, D.; Ouari, O.; et al. Detection of mitochondria-generated reactive oxygen species in cells using multiple probes and methods: Potentials, pitfalls, and the future. J. Biol. Chem. 2018, 293, 10363–10380. [Google Scholar] [CrossRef] [Green Version]
- Dikalov, S.I.; Polienko, Y.F.; Kirilyuk, I. Electron Paramagnetic Resonance Measurements of Reactive Oxygen Species by Cyclic Hydroxylamine Spin Probes. Antioxid. Redox Signal. 2018, 28, 1433–1443. [Google Scholar] [CrossRef]
- Strack, R.L.; Hein, B.; Bhattacharyya, D.; Hell, S.W.; Keenan, R.J.; Glick, B.S. A rapidly maturing far-red derivative of DsRed-Express2 for whole-cell labeling. Biochemistry 2009, 48, 8279–8281. [Google Scholar] [CrossRef]
- Li, X.; Zhao, X.; Fang, Y.; Jiang, X.; Duong, T.; Fan, C.; Huang, C.-C.; Kain, S.R. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998, 273, 34970–34975. [Google Scholar] [CrossRef] [Green Version]
- Froger, A.; Hall, J.E. Transformation of plasmid DNA into E. coli using the heat shock method. J. Vis. Exp. 2007, e253. [Google Scholar] [CrossRef]
- Kankaanpää, P.; Paavolainen, L.; Tiitta, S.; Karjalainen, M.; Päivärinne, J.; Nieminen, J.; Marjomäki, V.; Heino, J.; White, D.J. BioImageXD: An open, general-purpose and high-throughput image-processing platform. Nat. Methods 2012, 9, 683–689. [Google Scholar] [CrossRef]
- Yeo, W.S.; Lee, J.H.; Lee, K.C.; Roe, J.H. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 2006, 61, 206–218. [Google Scholar] [CrossRef]
- Lee, K.-C.; Yeo, W.-S.; Roe, J.-H. Oxidant-Responsive Induction of the suf Operon, Encoding a Fe-S Assembly System, through Fur and IscR in Escherichia coli. J. Bacteriol. 2008, 190, 8244–8247. [Google Scholar] [CrossRef] [Green Version]
- Fuangthong, M.; Jittawuttipoka, T.; Wisitkamol, R.; Romsang, A.; Duang-nkern, J.; Vattanaviboon, P.; Mongkolsuk, S. IscR plays a role in oxidative stress resistance and pathogenicity of a plant pathogen, Xanthomonas campestris. Microbiol. Res. 2015, 170, 139–146. [Google Scholar] [CrossRef]
- Chen, L.; Jiang, Z.G.; Khan, A.A.; Chishti, A.H.; McKnight, C.J. Dematin exhibits a natively unfolded core domain and an independently folded headpiece domain. Protein Sci. 2009, 18, 629–636. [Google Scholar] [CrossRef] [Green Version]
- Nagar, N.; Ecker, N.; Loewenthal, G.; Avram, O.; Ben-Meir, D.; Biran, D.; Ron, E.; Pupko, T. Harnessing machine learning to unravel protein degradation in Escherichia coli. Msystems 2021, 6, e01296-20. [Google Scholar] [CrossRef]
- Bujard, H.; Gentz, R.; Lanzer, M.; Stueber, D.; Mueller, M.; Ibrahimi, I.; Haeuptle, M.-T.; Dobberstein, B. [26] A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Methods Enzymol. 1987, 155, 416–433. [Google Scholar]
- Laboratories, B.R. BRL pUC host: E. coli DH5α competent cells. Focus 1986, 8, 9. [Google Scholar]
- Justino, M.C.; Almeida, C.C.; Teixeira, M.; Saraiva, L.M. Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J. Biol. Chem. 2007, 282, 10352–10359. [Google Scholar] [CrossRef] [Green Version]
- Vine, C.E.; Justino, M.C.; Saraiva, L.M.; Cole, J. Detection by whole genome microarrays of a spontaneous 126-gene deletion during construction of a ytfE mutant: Confirmation that a ytfE mutation results in loss of repair of iron-sulfur centres in proteins damaged by oxidative or nitrosative stress. J. Microbiol. Methods 2010, 81, 77–79. [Google Scholar] [CrossRef]
- Smirnova, G.V.; Zakirova, O.N.; Oktiabr’skiĭ, O.N. [Role of the antioxidant system in response of Escherichia coli bacteria to cold stress]. Mikrobiologiia 2001, 70, 55–60. [Google Scholar]
- Cardinale, S.; Joachimiak, M.P.; Arkin, A.P. Effects of genetic variation on the E. coli host-circuit interface. Cell Rep. 2013, 4, 231–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicol, M.J.; Brubaker, T.R.; Honish, B.J.; Simmons, A.N.; Kazemi, A.; Geissel, M.A.; Whalen, C.T.; Siedlecki, C.A.; Bilen, S.G.; Knecht, S.D. Antibacterial effects of low-temperature plasma generated by atmospheric-pressure plasma jet are mediated by reactive oxygen species. Sci. Rep. 2020, 10, 3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Choi, H. Single-Cell, Time-Lapse Reactive Oxygen Species Detection in E. coli. Curr. Protoc. Cell Biol. 2018, 80, e60. [Google Scholar] [CrossRef] [PubMed]
- Romsang, A.; Duang-Nkern, J.; Leesukon, P.; Saninjuk, K.; Vattanaviboon, P.; Mongkolsuk, S. The iron-sulphur cluster biosynthesis regulator IscR contributes to iron homeostasis and resistance to oxidants in Pseudomonas aeruginosa. PLoS ONE 2014, 9, e86763. [Google Scholar] [CrossRef]
- Izert, M.A.; Klimecka, M.M.; Górna, M.W. Applications of Bacterial Degrons and Degraders—Toward Targeted Protein Degradation in Bacteria. Front. Mol. Biosci. 2021, 8, 348. [Google Scholar] [CrossRef]
- Cameron, D.E.; Collins, J.J. Tunable protein degradation in bacteria. Nat. Biotechnol. 2014, 32, 1276–1281. [Google Scholar] [CrossRef]
- Moen, B.; Janbu, A.O.; Langsrud, S.; Langsrud, Ø.; Hobman, J.L.; Constantinidou, C.; Kohler, A.; Rudi, K. Global responses of Escherichia coli to adverse conditions determined by microarrays and FT-IR spectroscopy. Can. J. Microbiol. 2009, 55, 714–728. [Google Scholar] [CrossRef] [Green Version]


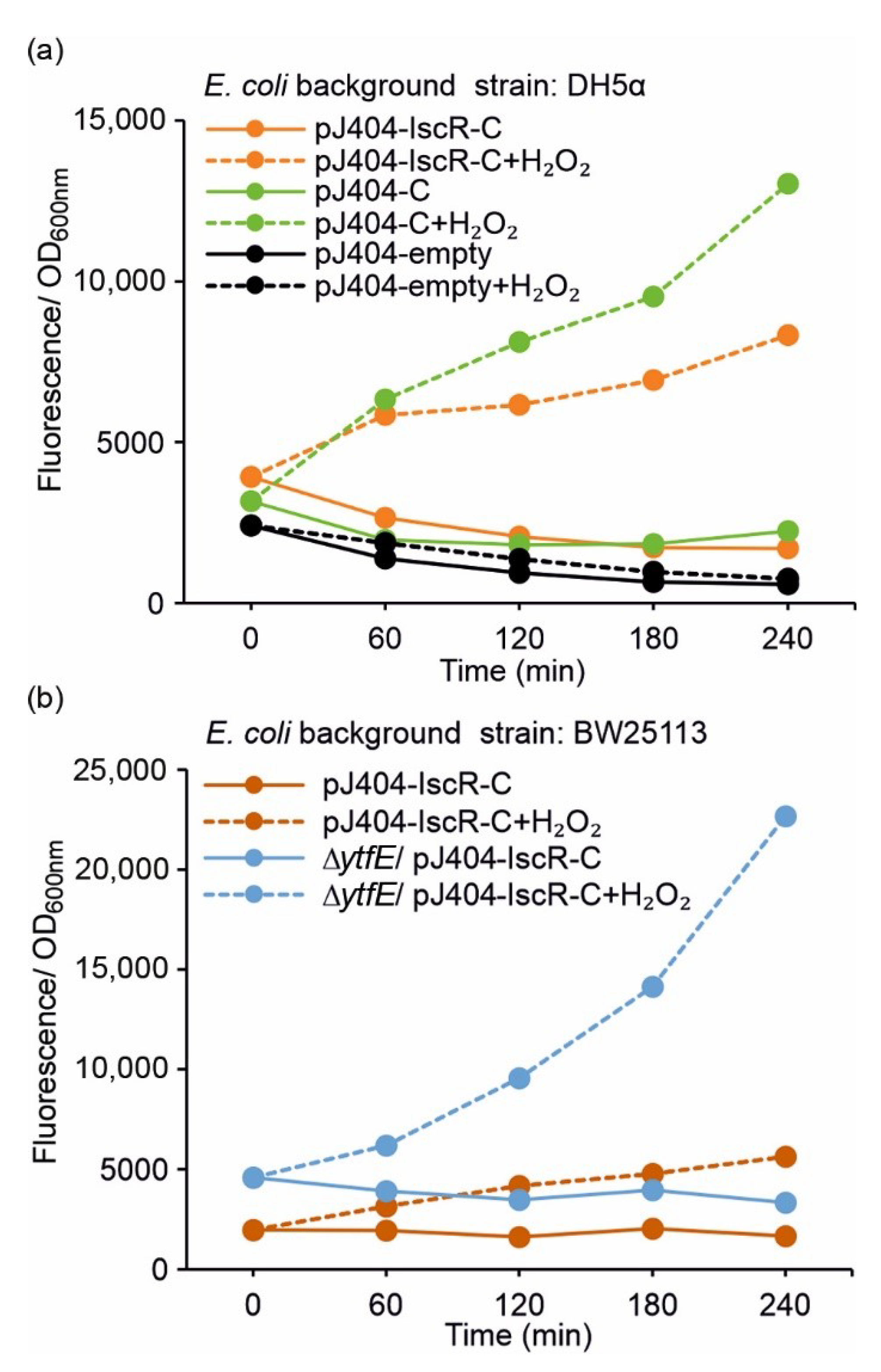




| Background E. coli Strain | Plasmid | Description |
|---|---|---|
| DH5α | pJ404-IscR-C | The default reporter system to monitor oxidative damage |
| DH5α | pJ404-C | Reference system without IscR overexpression |
| DH5α | pJ404 empty plasmid | Negative background control (fluorescence baseline) |
| BW25113 | pJ404-IscR-C | Reporter system in alternative host and Reference for BW25113 ∆ytfE |
| BW25113 ∆ytfE | pJ404-IscR-C | Reporter system in redox-sensitive host strain |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dandapani, H.; Kankaanpää, P.; Jones, P.R.; Kallio, P. A Plasmid-Based Fluorescence Reporter System for Monitoring Oxidative Damage in E. coli. Sensors 2022, 22, 6334. https://doi.org/10.3390/s22176334
Dandapani H, Kankaanpää P, Jones PR, Kallio P. A Plasmid-Based Fluorescence Reporter System for Monitoring Oxidative Damage in E. coli. Sensors. 2022; 22(17):6334. https://doi.org/10.3390/s22176334
Chicago/Turabian StyleDandapani, Hariharan, Pasi Kankaanpää, Patrik R. Jones, and Pauli Kallio. 2022. "A Plasmid-Based Fluorescence Reporter System for Monitoring Oxidative Damage in E. coli" Sensors 22, no. 17: 6334. https://doi.org/10.3390/s22176334
APA StyleDandapani, H., Kankaanpää, P., Jones, P. R., & Kallio, P. (2022). A Plasmid-Based Fluorescence Reporter System for Monitoring Oxidative Damage in E. coli. Sensors, 22(17), 6334. https://doi.org/10.3390/s22176334







