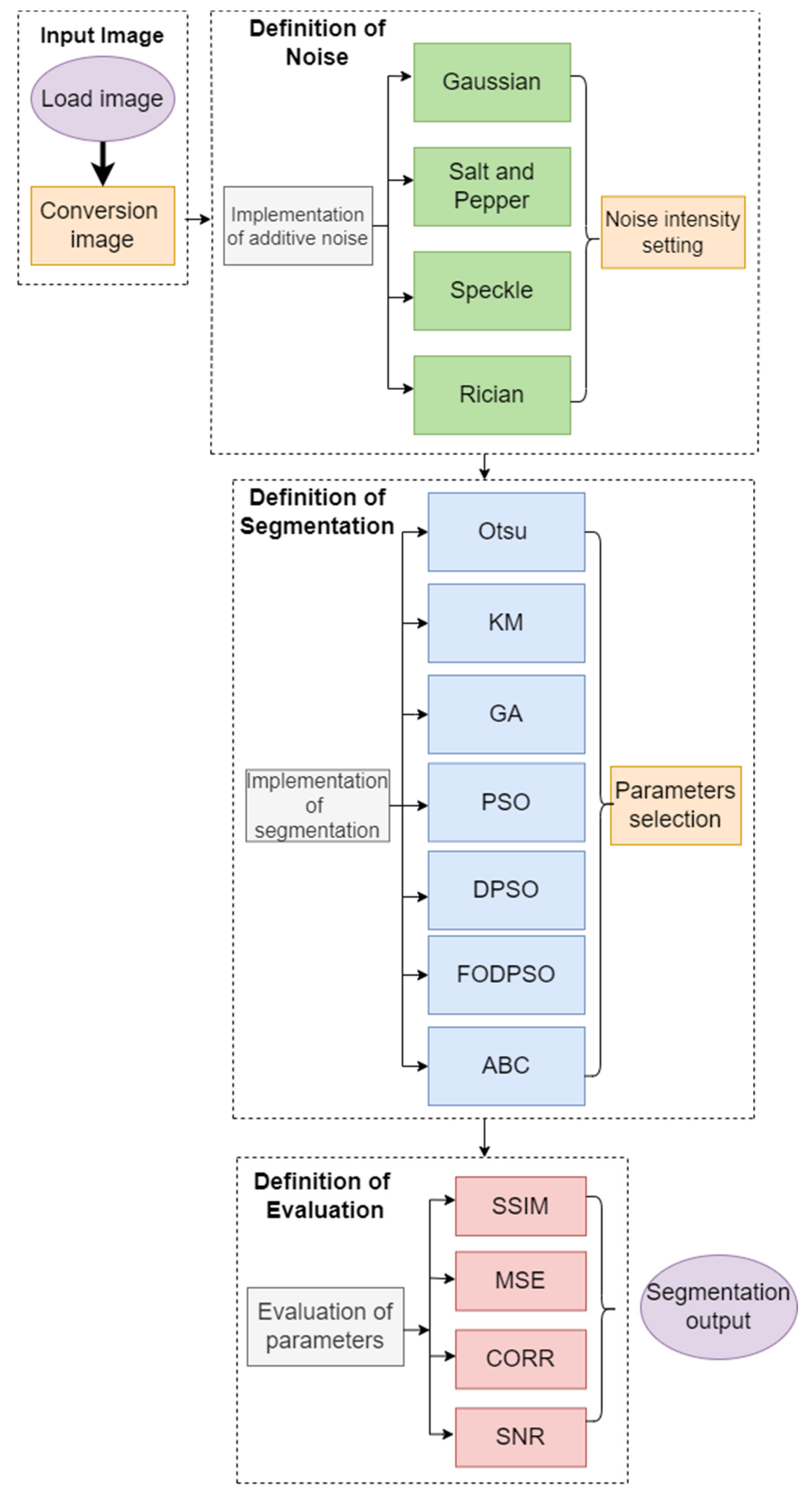
In this section, we introduce quantitative results of testing analyzed thresholding-based segmentation strategies. Here, we provide several types of characteristics to provide an objective view of the segmentation performance and limitations. We provide examples of graphical comparisons of the segmentation methods, which show the influence of the variable image noise of segmentation maps. For the generation of the segmentation maps, we use an artificial color coding. Where each single color represents one region of the segmentation model. To provide a complex view on the segmentation performance, we provide this testing for a variable number of segmentation classes because this parameter has a substantial effect on the segmentation performance. One of the important performance features is the time complexity, thus we provide time requirements of individual segmentation methods. This aspect is substantially important when performing a simultaneous segmentation of a stack of MR images. In order to show a statistical significance between the routine methods and optimized segmentation models, we provide the statistical testing of significance of p-values for median tests. The last quantitative analysis deals with the extraction of clinically important cartilage features including the area, perimeter, and cartilage skeleton. Here, we show differences of automatic segmentation and the gold standards. Lastly, we provide a presentation of the software environment, which integrates individual reported segmentation strategies with the possibility of selecting steering parameters of segmentation.
4.1. Quantitative Segmentation Evaluation
Gradual noise dynamics has a substantial effect on the pixel’s distribution as we mention in the previous examples. In our study, we utilize this fact to test the robustness of segmentation strategies to justify the impact of optimization elements for the performance of regional segmentation as we describe further.
The first analysis, which we provide is aimed on the graphical evaluation of the analyzed segmentation methods under gradual increasing noise influence. Based on such results, we can subjectively observe clearly visible notable differences in individual methods in segmentation maps. As the example, we provide the comparison (
Figure 10 and
Figure 11) for all the methods for salt and pepper noise with three levels of density: 0.1, 0.5, and 0.7 and Rician noise with three settings:
.
The segmentation results are interpreted in the form of segmentation maps in the color spectrum (
Figure 10 and
Figure 11). The interpretation of these color maps is that each segmentation region in the segmentation map is represented by a single value. Thus, the number of colors corresponds with the number of regions. Each such regional model can be interpreted as a transformation of the scale of intensity values to the number of regions. For instance, 8-bit images (256 intensity values) are transformed into four intensities (segmentation model with four regions). The main aim of this analysis is to objectively report how the distribution of a pixel’s assignment into individual regions are modified under the influence of additive noise against the gold standard (segmentation without additive noise influence).
Based on such experimental results, it is noticeable that an increasing noise intensity can significantly impair the quality of the segmentation results. For lower noise levels the segmentation results point out on a good performance, for example the ABC algorithm does not exhibit more significant signs of the noise. On the other hand, higher levels of noise cause significant impairment of the segmentation model consistency. In order to objectively justify this fact, we further provide a robust testing of these segmentation methods based on the mentioned evaluation parameters, to objectively show the change of segmentation performance among individual methods, and also how the number of regions influence the dynamic of segmentation performance. To better justify the testing scheme, we provide testing on routine approaches of Otsu thresholding and K-means clustering. Here, we only set the number of the segmentation regions. Contrarily, in the evolutionary strategies, including ABC, PSO, DPSO, and FPSO, we use a unified number of iterations, 100 (PSO1, GA1 and ABC1) and 500 (PSO2, GA2 and ABC2), and a population size of 50 (PSO1, GA1 and ABC1) and 200 (PSO2, GA2 and ABC2).
Here, we provide the quantitative comparison of individual optimization techniques for image thresholding-based regional segmentation against selected conventional segmentation approaches, including Otsu hard thresholding and K-means nonhierarchical clustering for regional segmentation. We provide dynamical feature extractions of these methods, reporting effectivity for each noise level and robustness in the form of the trend of the evaluation parameters upon additive noise with dynamic intensity, measured by the mean squared error (MSE), the index of correlation (CORR), the structural similarity index (SSIM), and the signal to noise ratio (SNR). As the example, we provide these characteristics (
Figure 12,
Figure 13,
Figure 14 and
Figure 15) for the segmentation models with four regions. The provided characteristics are constructed for 1000 images, where the results for each level of each noise are averaged.
Judging by the experimental results, significant differences in effectivity among individual methods are notable. The trends of the parameters of similarity (SSIM, CORR, and SNR) for Otsu and K-means exhibit significantly lower values when comparing with the evolutionary algorithms. That indicates the notable worse results of these routine algorithms in the comparison with the optimization techniques. The higher these parameters are, the better the performance of respective segmentation is achieved. On the other hand, these routine approaches from the view of MSE exhibit the most rapid increasing trend when comparing with optimization techniques. This is also a sign of the much worse effectivity of Otsu and K-means against the elements of artificial intelligence.
Besides these characteristics, we also publish the averaged results of individual parameters for individual methods. Note that the individual results are average for each parameter and the type of noise. These characteristics (
Table 3,
Table 4,
Table 5 and
Table 6) well reflect a global view on the respective segmentation performance. All these comparisons are provided for four segmentation classes. Red values represent the worst results for each test; contrarily, green values represent the best results for each test.
Based on the provided statistical results, we have an insight on a global behavior of the segmentation strategies. Judging by these experimental results, it is notable that the highest performance is achieved in most cases by the ABC algorithm. This is expected due to its enhanced segmentation strategy because it used the fuzzy soft thresholding instead of hard defined thresholds. The further expected fact is the methods with the lowest performance. Here, in most cases are Otsu and K-means, which are comparably worse than the genetic and evolutionary algorithms. The last interesting fact is that PSO and its variants achieve relatively similar results without more significant differences. It is notable that the SNR parameter reports relatively small values. This fact is caused by applying SNR on segmentation maps in the form of index matrixes (for each region we have one unique value) instead of intensity values, where we would have a distribution of 256 intensity levels (in the case of 8-bit images). The MSE parameter achieves small values as well. Here, it is expected, due to the nature of MSE. The lower the MSE is, the higher the agreement between the gold standard and the respective segmentation results we have.
Table 3,
Table 4,
Table 5 and
Table 6 provide the descriptive characteristics of averaged values from all the noise settings for the individual evaluation parameters (SSIM, MSE, CORR, and SNR). These results indicate a better performance of the segmentation strategies, which are optimized with evolutionary or genetic algorithms. On the other hand, these results do not report a statistical significance of testing. Therefore, we further publish statistical testing of the optimized methods against the conventional methods: Otsu thresholding and K-means clustering. Here, we aimed to provide the testing of statistical significance of the mean value for each optimized segmentation strategy against the methods Otsu and K-means for all the evaluation parameters and types of noise. Using paired
t-tests for mean values assumes that the tested data come from the normal distribution of probability. We tested all the distributions of SSIM, MSE, CORR, and SNR whether they come from the normal distribution. We used the null hypothesis (
) that the data come from the normal distribution and against the alternative hypothesis (
. We place the alternative hypothesis (
), which expresses that the data do not come from the normal distribution:
. For the testing, we use the chi-square test (
test) with the level of significance:
and the confidence interval:
. In all the tested parameters SSIM, MSE, CORR, and SNR for the noise: Gaussian, salt and pepper, speckle, and Rician, we found out that the
p-value is less than
(5%). Therefore, we reject the null hypothesis on the given significance level of 5% that the data come from the normal distribution. Since the data do not come from the normal distribution, we cannot use a paired
t-test, but we use statistical testing of the median with the Wilcoxon rank sum test. We provide testing for the same significance level:
and confidence interval:
. For all the median tests apart from the MSE, we define the null hypothesis as that the median value for the respective evaluation parameter for ABC, GA, PSO, DPSO, or FPSO is greater than the median for Otsu or K-means (
). In the case of MSE, we use the hypothesis:
, here, we supposed that a lower median represents less significant differences in segmentation effectivity. Here, (*) stands for the median value of the respective method and parameter,
A is the median of Otsu thresholding, and
B is the median of K-means. Against
, we put the alternative hypothesis as follows:
. In the
Table 7,
Table 8,
Table 9 and
Table 10, we present the
p-values for each test. For each parameter, we report two
p-values, where the first indicates the test against the Otsu method (
or
—for MSE) and the second against K-means (
or
—for MSE). The cases when we reject the null hypothesis
are indicated as red.
Statistical testing of significance based on the Wilcoxon rank sum test of comparing medians was supposed to declare a level of significance between the respective optimized segmentation strategy and Otsu- or K-means-based segmentation. In order to report this statistical significance, we provide the
p-values for each test, where its value declares the power of the test (
Table 7,
Table 8,
Table 9 and
Table 10). In the context of the employment of genetic algorithms in some cases, we reject the null hypothesis, which means the routine segmentation strategies achieved a significantly higher median. From the global view, in most cases, the evolutionary strategies achieved a higher median than routine algorithms (
). In these cases, we fail to reject the null hypothesis. The
p-value also enables measuring the power of the test. The higher it is, the more significant differences we achieved based on the test. In this context, in the case of the ABC algorithm, we normally achieved higher
p-values in the comparison with other methods. That shows that the combination of fuzzy thresholding with the ABC algorithm appears as the best segmentation strategy in this study.
In the last part of quantitative comparison, we compare variable numbers of segmentation regions to justify this effect on the segmentation performance. Here, we provide a comparison of 4, 7, and 10 regions of the ABC algorithm for Rician noise (
Figure 16). As it is obvious, the number of regions plays an important role in the context of segmentation performance. Based on these results, we can objectively conclude that the segmentation with a lover number of regions mostly achieves a better segmentation performance, contrarily, 10 segmentation regions exhibit the least segmentation performance. The last comparison (
Figure 17) that we provide is a detailed insight on the performance of the ABC algorithm with two various settings (ABC
1, ABC
2) as we indicated earlier. Here, we show the comparison of these two ABC alternatives for 4, 7, and 10 regions. Furthermore, here, we can observe significant differences in the algorithm’s performance. Mostly, the higher number of regions are set, the worse segmentation results we achieve.
Besides the quantitative characteristics in this section, we also publish a comparison of time requirements. We provide testing (
Table 11 and
Table 12) of the time complexity for each analyzed method for the simultaneous computing of 100 MR images of articular cartilage (same images were used for all the methods). We provide testing on the following hardware configuration: Intel(R) Core(TM) i5-10300H CPU@2.50 GHz, RAM: 16.0 GB. Based on the achieved results, the significant differences among the methods are notable. Otsu and K-means have the lowest time complexity in all the cases. This is expected due to not requiring optimization techniques. Thus, the main benefit of these methods would be their speed. On the other hand, the use of genetic and optimization algorithms is time demanding, as we declare in the following tables. By this comparison, the slowest method appears to be the genetic algorithm.
4.3. Clinical Important Features Extraction of Articular Cartilage
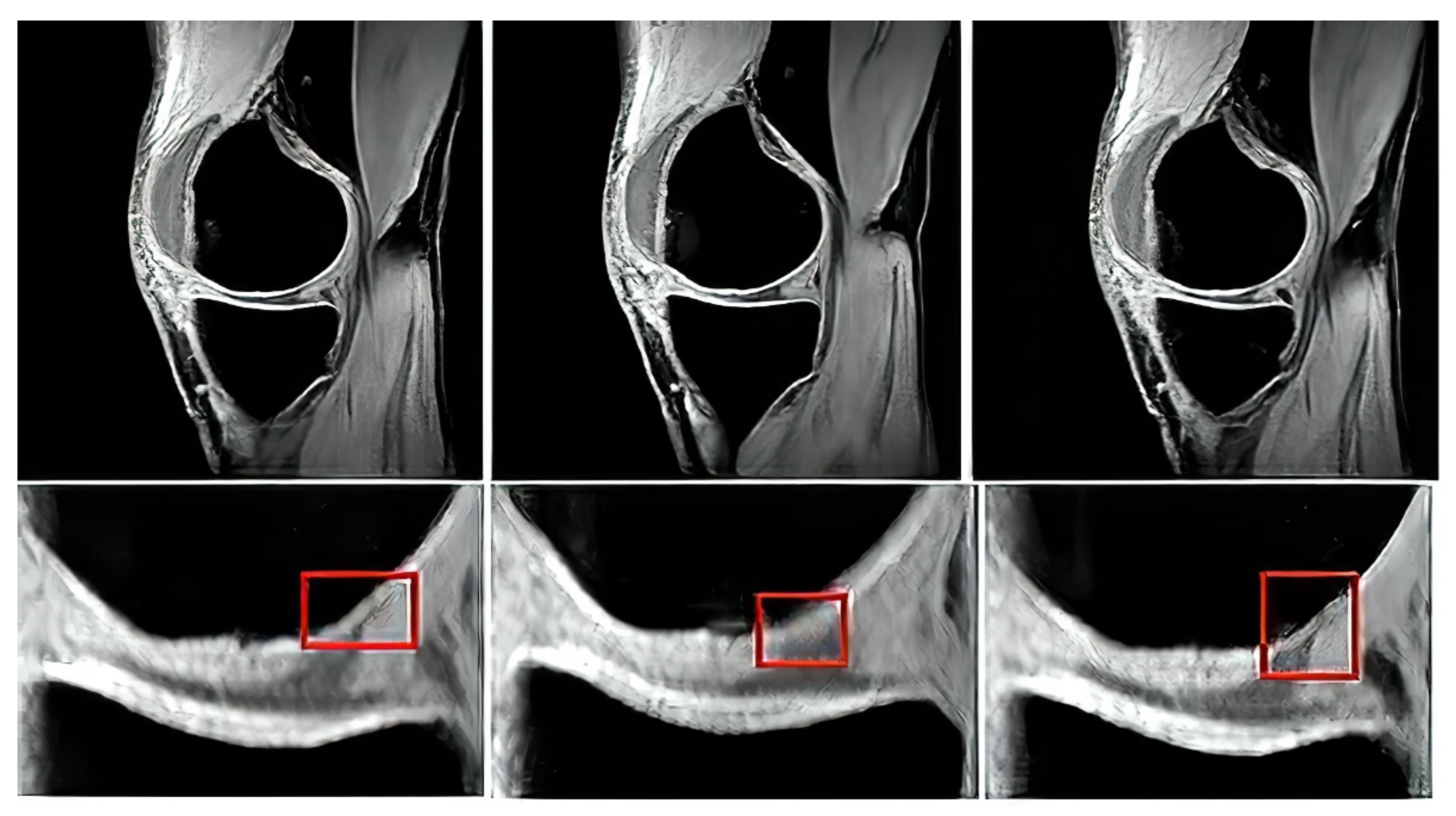
Based on the reported analysis of the segmentation performance, mostly the combination of fuzzy thresholding with the ABC evolutionary algorithms appeared as the best segmentation strategy, judging by reported objectivization parameters and mainly provided statistical tests of significance. In this subsection, we would like to provide the last analysis of selected features extraction of articular cartilage from MR images based on the fuzzy thresholding with the ABC algorithm. The aim of this analysis is firstly computing a multiregional segmentation model, allowing for a decomposition of the MR image into a finite number (in this case five) segmentation regions. Consequently, a region, representing the articular cartilage, is selected (
Figure 20) as the region of interest, while the rest of the segmentation regions are suppressed from the segmentation model (
Figure 20). By this selection scheme, we obtain a binary segmentation model, exclusively classifying the articular cartilage from the rest of the tissues in the MR images.
Figure 20 also presents a multiregional segmentation of a part of articular cartilage (femoral cartilage) affected by osteoarthritis of I. grade, which is notable by two segmented lobes of the articular cartilage, and between them is a gap, where the cartilage is missing. To objectize the quality of the articular cartilage extraction and the preciseness of the reported features, we extracted the same features for the gold standard manual segmentation of articular cartilage. Consequently, the feature differences are compared to quantify the segmentation effectivity of articular cartilage detection. Note that we used the following settings for the ABC algorithm: 100 iterations and population size 50. The following cartilage features are considered for evaluation:
Cartilage area—a total count of the pixels, belonging to the model of articular cartilage.
Cartilage perimeter—a perimeter of the cartilage model. Here, we used Sobel edge operator for the detection of cartilage borders, and consequently counted the border pixels.
Skeleton of cartilage—the detection of cartilage skeleton and computing its length.
Figure 20.
Example of segmentation results for articular cartilage and its features extraction based on fuzzy thresholding with ABC optimization: (a) gold standards by manual annotation, (b) binary segmentation, (c) native MR image with area of interest indicated by the green square (top) and multiregional segmentation with 4 regions (bottom), where yellow contours reflect two lobes of articular cartilage from region of interest, and (d) binary extraction of articular cartilage fused with the gold standard (red contour).
Figure 20.
Example of segmentation results for articular cartilage and its features extraction based on fuzzy thresholding with ABC optimization: (a) gold standards by manual annotation, (b) binary segmentation, (c) native MR image with area of interest indicated by the green square (top) and multiregional segmentation with 4 regions (bottom), where yellow contours reflect two lobes of articular cartilage from region of interest, and (d) binary extraction of articular cartilage fused with the gold standard (red contour).
Based on the segmentation form as binary images, representing extracted articular cartilage and its respective features, we compute descriptive statistics, pointing out on individual distribution’s error functions, which show percentual differences of individual features, and a distribution of values for the individual parameters of segmentation performance (SSIM and index of correlation). Here, the evaluation parameters were computed between the gold standard binary image and the results of the fuzzy thresholding with the ABC algorithm.
Figure 21 provides a graphical representation of the distributions of differences for cartilage features and the distribution of values for performance parameters for fuzzy soft thresholding with ABC optimization.
Based on the results of the quantitative analysis of difference function for the extracted features, we did not achieve significant differences between the gold standard images and fuzzy soft thresholding with the ABC algorithm. Mostly the distributions of difference function are kept under 6% of difference. Based on this analysis, we provide the descriptive characteristics (
Table 13), which reports the median and standard deviation for each parameter. Based on these results, the best result in median difference is achieved for the feature of skeleton length (2.42%); contrarily, the worst median difference is achieved for the area (4.12%). From the view of measuring variability (standard deviation) of the difference function, the lowest difference is achieved for the skeleton (1.38%) in the contrast with the parameter area, where the difference was the worst (2.44%). The second studied aspect is the performance parameters: the index of correlation and the SSIM. Here, we achieved a higher median for correlation (0.94), where the median for the SSIM was 0.89. Furthermore, from the view of standard deviation, representing the concentration of values is better than the index of correlation (0.017), while in SSIM we achieved 0.028.