Automated Detection of Sleep Apnea-Hypopnea Events Based on 60 GHz Frequency-Modulated Continuous-Wave Radar Using Convolutional Recurrent Neural Networks: A Preliminary Report of a Prospective Cohort Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Sleep Questionnaires and Polysomnography
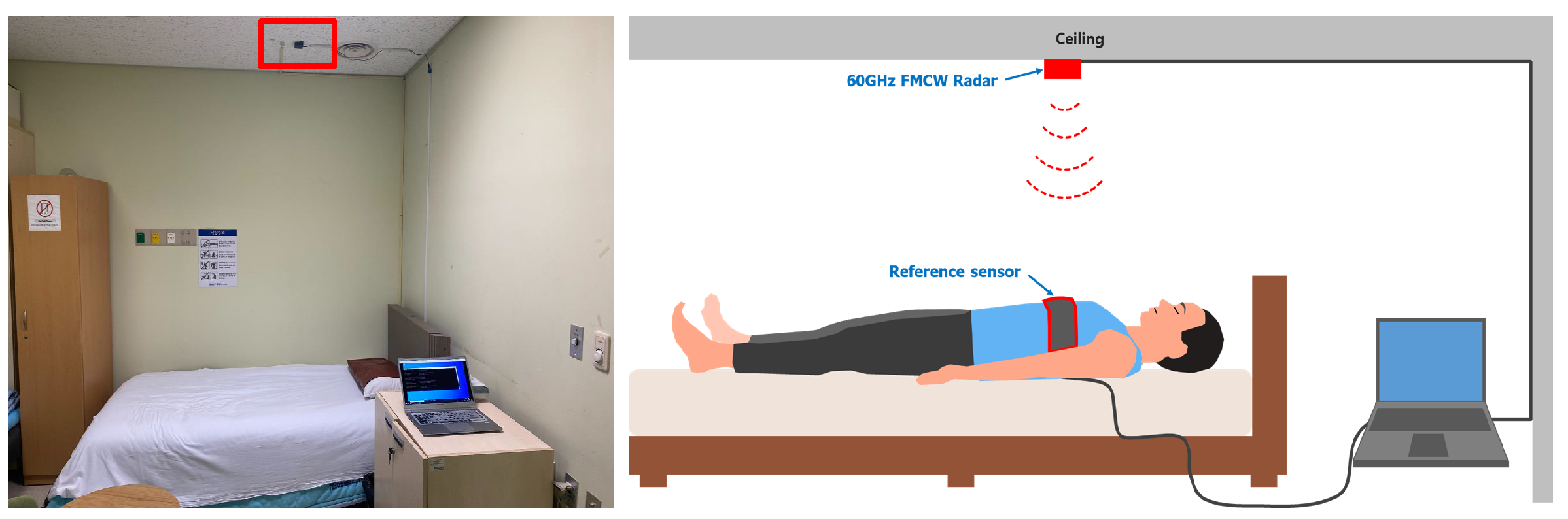
2.3. Radar Setup
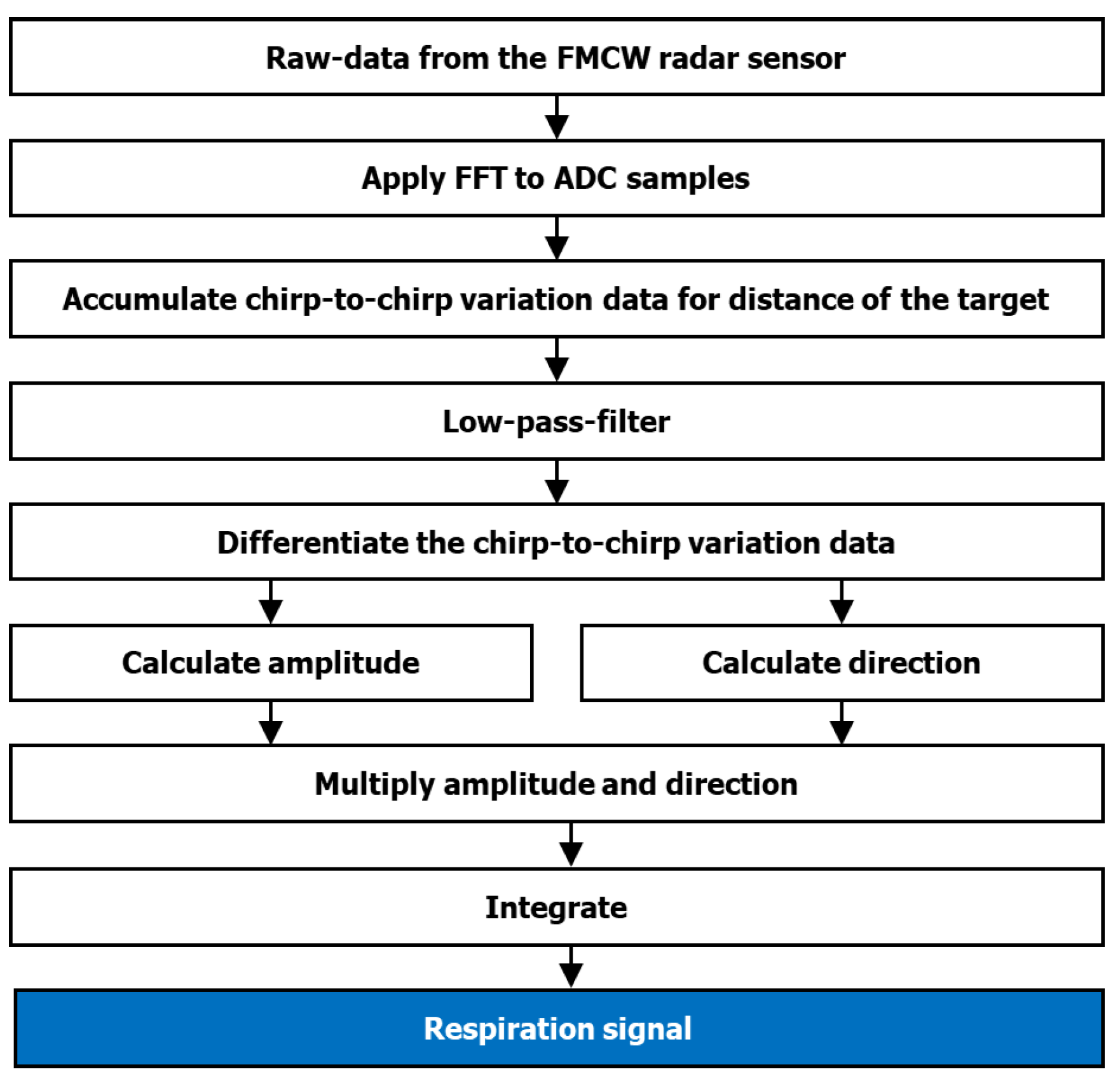
2.4. Data Preparation and Preprocessing
2.5. Model Development
2.6. Performance Evaluation
3. Results
3.1. Study Participants
3.2. Per-Segment Classification Performance
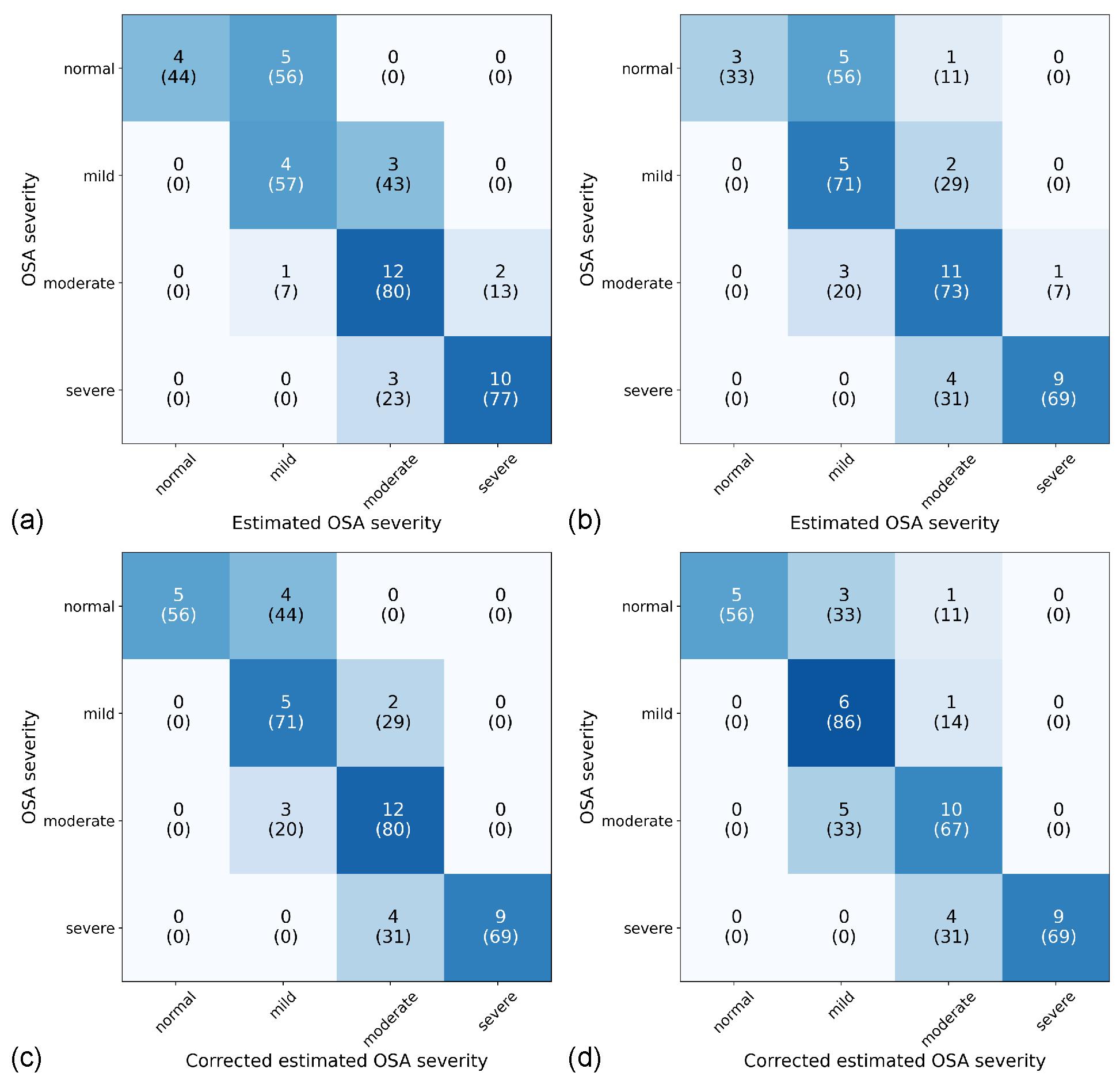
3.3. Global Event Detection Performance
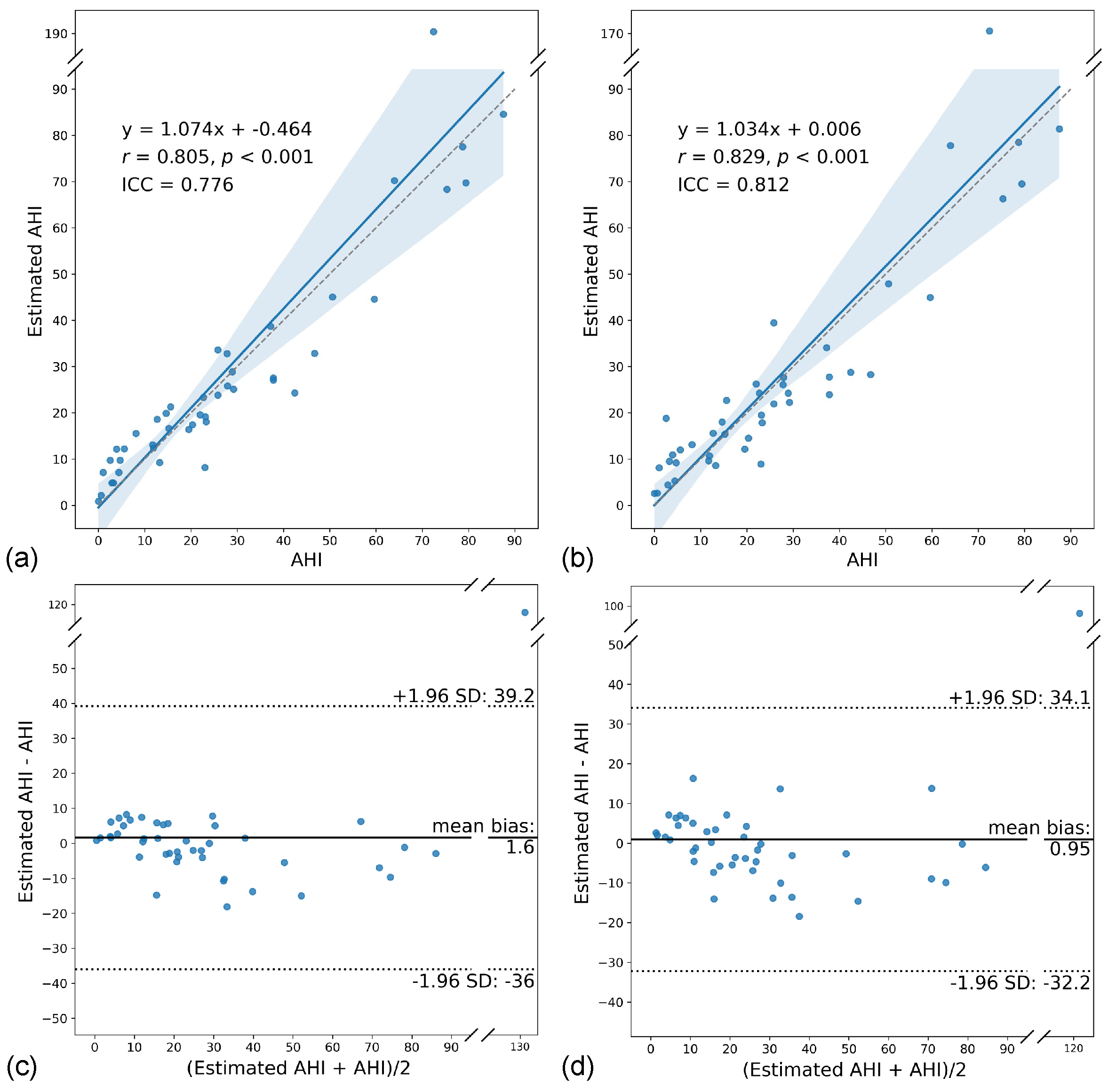
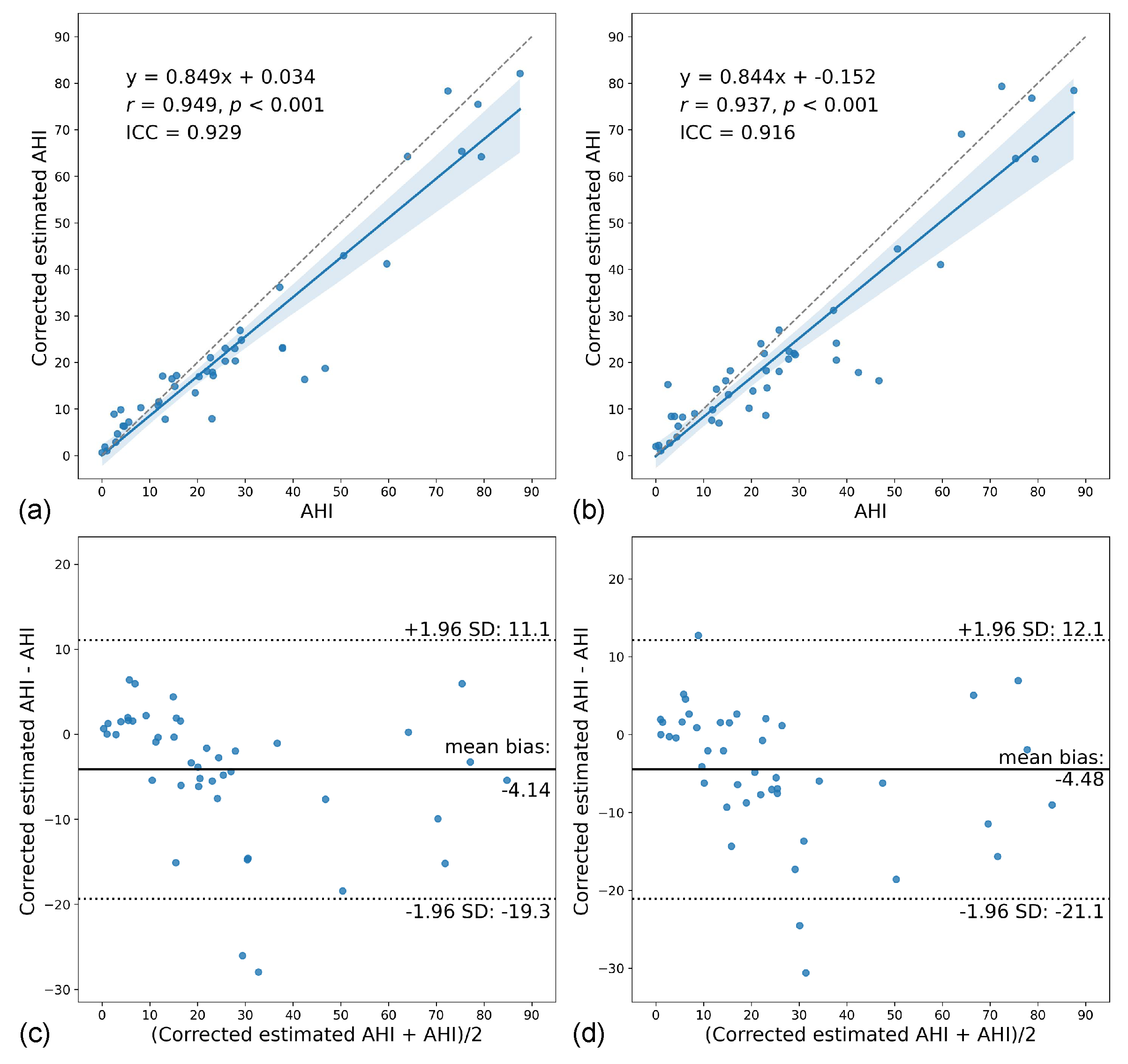
3.4. AHI Estimation Performance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olson, E.J.; Moore, W.R.; Morgenthaler, T.I.; Gay, P.C.; Staats, B.A. Obstructive sleep apnea-hypopnea syndrome. Mayo Clin. Proc. 2003, 78, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Young, T.; Palta, M.; Dempsey, J.; Skatrud, J.; Weber, S.; Badr, S. The occurrence of sleep-disordered breathing among middle-aged adults. N. Engl. J. Med. 1993, 328, 1230–1235. [Google Scholar] [CrossRef] [PubMed]
- Koo, D.L.; Nam, H.; Thomas, R.J.; Yun, C.H. Sleep Disturbances as a Risk Factor for Stroke. J. Stroke Cerebrovasc. Dis. 2018, 20, 12–32. [Google Scholar] [CrossRef] [PubMed]
- Andrade, A.G.; Bubu, O.M.; Varga, A.W.; Osorio, R.S. The Relationship between Obstructive Sleep Apnea and Alzheimer’s Disease. J. Alzheimers. Dis. 2018, 64, S255–S270. [Google Scholar] [CrossRef]
- Somers, V.K.; White, D.P.; Amin, R.; Abraham, W.T.; Costa, F.; Culebras, A.; Daniels, S.; Floras, J.S.; Hunt, C.E.; Olson, L.J.; et al. Sleep apnea and cardiovascular disease: An American Heart Association/american College of Cardiology Foundation Scientific Statement from the American Heart Association Council for High Blood Pressure Research Professional Education Committee, Council on Clinical Cardiology, Stroke Council, and Council On Cardiovascular Nursing. In collaboration with the National Heart, Lung, and Blood Institute National Center on Sleep Disorders Research (National Institutes of Health). Circulation 2008, 118, 1080–1111. [Google Scholar] [CrossRef]
- Kapur, V.K.; Auckley, D.H.; Chowdhuri, S.; Kuhlmann, D.C.; Mehra, R.; Ramar, K.; Harrod, C.G. Clinical Practice Guideline for Diagnostic Testing for Adult Obstructive Sleep Apnea: An American Academy of Sleep Medicine Clinical Practice Guideline. J. Clin. Sleep Med. 2017, 13, 479–504. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.Y.; Song, K.; Shin, K.; Sohn, J.; Cho, S.H.; Chang, J.H. Noncontact Sleep Study by Multi-Modal Sensor Fusion. Sensors 2017, 17, 1685. [Google Scholar] [CrossRef]
- Le Bon, O.; Staner, L.; Hoffmann, G.; Dramaix, M.; San Sebastian, I.; Murphy, J.R.; Kentos, M.; Pelc, I.; Linkowski, P. The first-night effect may last more than one night. J. Psychiatr. Res. 2001, 35, 165–172. [Google Scholar] [CrossRef]
- Newell, J.; Mairesse, O.; Verbanck, P.; Neu, D. Is a one-night stay in the lab really enough to conclude? First-night effect and night-to-night variability in polysomnographic recordings among different clinical population samples. Psychiatry Res. 2012, 200, 795–801. [Google Scholar] [CrossRef]
- Matar, G.; Lina, J.M.; Carrier, J.; Kaddoum, G. Unobtrusive sleep monitoring using cardiac, breathing and movements activities: An exhaustive review. IEEE Access 2018, 6, 45129–45152. [Google Scholar] [CrossRef]
- Addison, A.P.; Addison, P.S.; Smit, P.; Jacquel, D.; Borg, U.R. Noncontact Respiratory Monitoring Using Depth Sensing Cameras: A Review of Current Literature. Sensors 2021, 21, 1135. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.P.; Al-Jumaily, A.A.; Islam, S.M.S. Doppler radar-based non-contact health monitoring for obstructive sleep apnea diagnosis: A comprehensive review. Big Data Cogn. Comput. 2019, 3, 3. [Google Scholar] [CrossRef]
- Kebe, M.; Gadhafi, R.; Mohammad, B.; Sanduleanu, M.; Saleh, H.; Al-Qutayri, M. Human Vital Signs Detection Methods and Potential Using Radars: A Review. Sensors 2020, 20, 1454. [Google Scholar] [CrossRef]
- Vasireddy, R.; Roth, C.; Mathis, J.; Goette, J.; Jacomet, M.; Vogt, A. K-band Doppler radar for contact-less overnight sleep marker assessment: A pilot validation study. J. Clin. Monit. Comput. 2018, 32, 729–740. [Google Scholar] [CrossRef]
- Lee, Y.S.; Pathirana, P.N.; Steinfort, C.L.; Caelli, T. Monitoring and Analysis of Respiratory Patterns Using Microwave Doppler Radar. IEEE J. Transl. Eng. Health Med. 2014, 2, 1800912. [Google Scholar] [CrossRef]
- Kang, S.; Lee, Y.; Lim, Y.H.; Park, H.K.; Cho, S.H.; Cho, S.H. Validation of noncontact cardiorespiratory monitoring using impulse-radio ultra-wideband radar against nocturnal polysomnography. Sleep Breath. 2020, 24, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Al-Naji, A.; Gibson, K.; Lee, S.H.; Chahl, J. Monitoring of cardiorespiratory signal: Principles of remote measurements and review of methods. IEEE Access 2017, 5, 15776–15790. [Google Scholar] [CrossRef]
- Zhou, Y.; Shu, D.; Xu, H.; Qiu, Y.; Zhou, P.; Ruan, W.; Qin, G.; Jin, J.; Zhu, H.; Ying, K.; et al. Validation of novel automatic ultra-wideband radar for sleep apnea detection. J. Thorac. Dis. 2020, 12, 1286–1295. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, M.; Tojima, H.; Matsui, T. Non-contact diagnostic system for sleep apnea-hypopnea syndrome based on amplitude and phase analysis of thoracic and abdominal Doppler radars. Med. Biol. Eng. Comput. 2016, 54, 789–798. [Google Scholar] [CrossRef]
- Kang, S.; Kim, D.K.; Lee, Y.; Lim, Y.H.; Park, H.K.; Cho, S.H.; Cho, S.H. Non-contact diagnosis of obstructive sleep apnea using impulse-radio ultra-wideband radar. Sci. Rep. 2020, 10, 5261. [Google Scholar] [CrossRef]
- Javaid, A.Q.; Noble, C.M.; Rosenberg, R.; Weitnauer, M.A. Towards sleep apnea screening with an under-the-mattress IR-UWB radar using machine learning. In Proceedings of the 2015 IEEE 14th International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 9–11 December 2015. [Google Scholar] [CrossRef]
- Koda, T.; Sakamoto, T.; Okumura, S.; Taki, H.; Hamada, S.; Chin, K. Radar-based automatic detection of sleep apnea using support vector machine. In Proceedings of the 2020 International Symposium on Antennas and Propagation (ISAP), Osaka, Japan, 25–28 January 2021. [Google Scholar] [CrossRef]
- Zhao, H.; Hong, H.; Miao, D.; Li, Y.; Zhang, H.; Zhang, Y.; Li, C.; Zhu, X. A Noncontact Breathing Disorder Recognition System Using 2.4-GHz Digital-IF Doppler Radar. IEEE J. Biomed. Health Inform. 2019, 23, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Längkvist, M.; Karlsson, L.; Loutfi, A. A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recognit. Lett. 2014, 42, 11–24. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, N.; Swaminathan, R.; Deserno, T.M. Deep Learning on 1-D Biosignals: A Taxonomy-based Survey. Yearb. Med. Inform. 2018, 27, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Bai, X.; Yao, C. An end-to-end trainable neural network for image-based sequence recognition and its application to scene text recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 2298–2304. [Google Scholar] [CrossRef] [PubMed]
- Adavanne, S.; Politis, A.; Nikunen, J.; Virtanen, T. Sound event localization and detection of overlapping sources using convolutional recurrent neural networks. IEEE J. Sel. Top. Signal Process. 2019, 13, 34–48. [Google Scholar] [CrossRef]
- Cho, Y.W.; Lee, J.H.; Son, H.K.; Lee, S.H.; Shin, C.; Johns, M.W. The reliability and validity of the Korean version of the Epworth sleepiness scale. Sleep Breath. 2011, 15, 377–384. [Google Scholar] [CrossRef]
- Hoddes, E.; Zarcone, V.; Smythe, H.; Phillips, R.; Dement, W.C. Quantification of sleepiness: A new approach. Psychophysiology 1973, 10, 431–436. [Google Scholar] [CrossRef]
- Buysse, D.J.; Reynolds, C.F.; Monk, T.H.; Berman, S.R.; Kupfer, D.J. The Pittsburgh sleep quality index: A new instrument for psychiatric practice and research. Psychiatry Res. 1989, 28, 193–213. [Google Scholar] [CrossRef]
- Berry, R.B.; Budhiraja, R.; Gottlieb, D.J.; Gozal, D.; Iber, C.; Kapur, V.K.; Marcus, C.L.; Mehra, R.; Parthasarathy, S.; Quan, S.F.; et al. Rules for scoring respiratory events in sleep: Update of the 2007 AASM Manual for the Scoring of Sleep and Associated Events. Deliberations of the Sleep Apnea Definitions Task Force of the American Academy of Sleep Medicine. J. Clin. Sleep Med. 2012, 8, 597–619. [Google Scholar] [CrossRef] [PubMed]
- Park, B.K.; Boric-Lubecke, O.; Lubecke, V.M. Arctangent demodulation with DC offset compensation in quadrature Doppler radar receiver systems. IEEE Trans. Microw. Theory Tech. 2007, 55, 1073–1079. [Google Scholar] [CrossRef]
- Noguchi, H.; Kubo, H.; Mori, T.; Sato, T.; Sanada, H. Signal phase estimation for measurement of respiration waveform using a microwave Doppler sensor. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2013, 2013, 6740–6743. [Google Scholar] [CrossRef]
- Ma, J.; Chen, J.; Ng, M.; Huang, R.; Li, Y.; Li, C.; Yang, X.; Martel, A.L. Loss odyssey in medical image segmentation. Med. Image Anal. 2021, 71, 102035. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Youden, W.J. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Lipton, Z.C.; Elkan, C.; Naryanaswamy, B. Optimal Thresholding of Classifiers to Maximize F1 Measure. Mach. Learn. Knowl. Discov. Databases 2014, 8725, 225–239. [Google Scholar] [CrossRef]
- Haidar, R.; McCloskey, S.; Koprinska, I.; Jeffries, B. Convolutional neural networks on multiple respiratory channels to detect hypopnea and obstructive apnea events. In Proceedings of the 2018 International Joint Conference on Neural Networks (IJCNN), Rio de Janeiro, Brazil, 8–13 July 2018. [Google Scholar] [CrossRef]
- Van Steenkiste, T.; Groenendaal, W.; Deschrijver, D.; Dhaene, T. Automated Sleep Apnea Detection in Raw Respiratory Signals Using Long Short-Term Memory Neural Networks. IEEE J. Biomed. Health Inform. 2019, 23, 2354–2364. [Google Scholar] [CrossRef]
- Nassi, T.E.; Ganglberger, W.; Sun, H.; Bucklin, A.A.; Biswal, S.; van Putten, M.J.A.M.; Thomas, R.J.; Westover, M.B. Automated respiratory event detection using deep neural networks. arXiv 2021, arXiv:2101.04635. [Google Scholar]
- Kwon, H.B.; Son, D.; Lee, D.; Yoon, H.; Lee, M.H.; Lee, Y.J.; Choi, S.H.; Park, K.S. Hybrid CNN-LSTM network for real-time apnea-hypopnea event detection based on IR-UWB radar. IEEE Access 2022, 10, 17556–17564. [Google Scholar] [CrossRef]
- Mostafa, S.S.; Mendonça, F.; Ravelo-García, A.G.; Morgado-Dias, F. A Systematic Review of Detecting Sleep Apnea Using Deep Learning. Sensors 2019, 19, 4934. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Yoon, H.; Kim, H.S.; Kim, H.B.; Kwon, H.B.; Oh, S.M.; Lee, Y.J.; Park, K.S. Real-time apnea-hypopnea event detection during sleep by convolutional neural networks. Comput. Biol. Med. 2018, 100, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Motiian, S.; Piccirilli, M.; Adjeroh, D.A.; Doretto, G. Unified deep supervised domain adaptation and generalization. arXiv 2017, arXiv:1709.10190. [Google Scholar]
- Eiseman, N.A.; Westover, M.B.; Ellenbogen, J.M.; Bianchi, M.T. The impact of body posture and sleep stages on sleep apnea severity in adults. J. Clin. Sleep Med. 2012, 8, 655–666. [Google Scholar] [CrossRef]





| Variables | Normal | Mild OSA | Moderate OSA | Severe OSA | |
|---|---|---|---|---|---|
| Subject characteristics | Number (male/female) | 9 (2/7) | 7 (4/3) | 15 (10/5) | 13 (9/4) |
| Age | 46.4 ± 17.9 | 49.9 ± 18.0 | 57.9 ± 9.5 | 55.3 ± 17.5 | |
| Body mass index (kg/m2) | 24.6 ± 4.1 | 24.9 ± 1.9 | 25.2 ± 2.9 | 26.7 ± 17.5 | |
| Neck circumference (cm) | 36.7 ± 3.0 | 39.0 ± 3.6 | 38.6 ± 4.2 | 40.8 ± 3.1 | |
| ESS score | 4.8 ± 3.1 | 5.0 ± 3.3 | 9.0 ± 3.7 | 8.5 ± 3.8 | |
| SSS score | 2.2 ± 1.0 | 3.0 ± 0.8 | 2.0 ± 0.5 | 2.4 ± 0.8 | |
| PSQI score | 8.6 ± 3.2 | 11.0 ± 2.8 | 10.0 ± 3.5 | 9.7 ± 4.0 | |
| Polysomnographic data | Time in bed (min) | 362.6 ± 106.5 | 412.0 ± 16.2 | 396.0 ± 23.9 | 400.1 ± 24.4 |
| Total sleep time (min) | 294.4 ± 97.2 | 311.0 ± 43.6 | 314.8 ± 66.7 | 266.5 ± 90.6 | |
| Sleep latency (min) | 18.2 ± 30.5 | 17 ± 12.7 | 16.4 ± 27.6 | 13.8 ± 14.8 | |
| Sleep efficiency (%) | 80.1 ± 8.7 | 75.7 ± 11.7 | 78.9 ± 13.7 | 66.3 ± 22.1 | |
| N1 (%) | 12.3 ± 7.2 | 14.0 ± 2.6 | 16.9 ± 8.0 | 33.4 ±13.5 | |
| N2 (%) | 52.3 ± 5.0 | 51.4 ± 5.7 | 51.3 ± 6.4 | 47.2 ± 9.6 | |
| N3 (%) | 20.0 ± 3.0 | 19.2 ± 2.1 | 16.7 ± 5.3 | 8.5 ± 8.4 | |
| REM (%) | 15.4 ± 9.6 | 15.4 ± 6.7 | 15.0 ± 6.2 | 10.9 ± 7.9 | |
| Apnea index (events/h) | 0 | 0.1 ± 0.2 | 2.8 ± 3.2 | 20.1 ± 20.6 | |
| Hypopnea index (events/h) | 2.5 ± 1.9 | 10.3 ± 2.6 | 20.3 ± 4.2 | 34.6 ± 11.0 | |
| AHI (events/h) | 2.6 ± 1.7 | 11.1 ± 3.1 | 23.3 ± 4.4 | 59.2 ± 18.2 | |
| RERA index (events/h) | 0.1 ± 0.3 | 0 | 0 | 0 | |
| Arousal index (events/h) | 19.9 ± 10.7 | 20.8 ± 5.9 | 29.4 ± 10.4 | 53.9 ± 17.6 | |
| Lowest O2 saturation (%) | 89.0 ± 3.3 | 87.7 ± 2.6 | 83.6 ± 3.4 | 72.8 ± 11.2 | |
| Number of segments | Abnormal | 296 | 906 | 3917 | 6016 |
| Normal | 6298 | 4911 | 8063 | 4485 | |
| Model | Metric | Overall | Normal | Mild | Moderate | Severe |
|---|---|---|---|---|---|---|
| Binary | AUROC | 0.846 [0.842, 0.851] | 0.819 [0.793, 0.846] | 0.796 [0.780, 0.812] | 0.798 [0.789, 0.807] | 0.859 [0.852, 0.866] |
| Sensitivity | 0.744 (8289/11135) [0.736, 0.753] | 0.625 (185/296) [0.570, 0.680] | 0.657 (595/906) [0.626, 0.688] | 0.674 (2641/3917) [0.660, 0.689] | 0.809 (4868/6016) [0.799, 0.819] | |
| Specificity | 0.803 (19065/23757) [0.797, 0.808] | 0.868 (5464/6298) [0.859, 0.876] | 0.767 (3916/4911) [0.786, 0.809] | 0.781 (6293/8063) [0.771, 0.790] | 0.756 (3392/4485) [0.744, 0.769] | |
| PPV | 0.639 (8289/12981) [0.630, 0.648] | 0.182 (185/1019) [0.138, 0.226] | 0.374 (595/1590) [0.343, 0.406] | 0.599 (2641/4411) [0.583, 0.614] | 0.817 (4868/5961) [0.807, 0.826] | |
| NPV | 0.870 (19065/21911) [0.866, 0.874] | 0.980 (5464/5575) [0.977, 0.984] | 0.926 (3916/4227) [0.919, 0.934] | 0.831 (6293/7569) [0.823, 0.840] | 0.747 (3392/4540) [0.734, 0.760] | |
| Accuracy | 0.784 (27354/34892) [0.780, 0.788] | 0.857 (5649/6594) [0.848, 0.865] | 0.776 (4511/5817) [0.765, 0.786] | 0.746 (8934/11980) [0.738, 0.754] | 0.787 (8260/10501) [0.779, 0.794] | |
| Multiclass | AUROC | 0.844 [0.840, 0.849] | 0.816 [0.788, 0.843] | 0.807 [0.791, 0.822] | 0.795 [0.787, 0.804] | 0.858 [0.850, 0.865] |
| Sensitivity | 0.721 (8024/11135) [0.712, 0.729] | 0.628 (186/296) [0.573, 0.683] | 0.605 (548/906) [0.573, 0.637] | 0.638 (2498/3917) [0.623, 0.653] | 0.797 (4792/6016) [0.786, 0.807] | |
| Specificity | 0.813 (19305/23757) [0.808, 0.818] | 0.850 (5352/6298) [0.841, 0.859] | 0.829 (4070/4911) [0.818, 0.839] | 0.800 (6454/8063) [0.792, 0.809] | 0.765 (3429/4485) [0.752, 0.777] | |
| PPV | 0.643 (8024/12476) [0.634, 0.652] | 0.164 (186/1132) [0.122, 0.207] | 0.395 (548/1389) [0.363, 0.426] | 0.608 (2498/4107) [0.593, 0.624] | 0.819 (4792/5848) [0.810, 0.829] | |
| NPV | 0.861 (19305/22416) [0.857, 0.866] | 0.980 (5352/5462) [0.976, 0.983] | 0.919 (4070/4428) [0.912, 0.927] | 0.820 (6454/7873) [0.811, 0.828] | 0.737 (3429/4653) [0.724, 0.750] | |
| Accuracy | 0.783 (27329/34892) [0.779, 0.788] | 0.840 (5538/6594) [0.831, 0.849] | 0.794 (4618/5817) [0.783, 0.804] | 0.747 (8952/11980) [0.739, 0.755] | 0.783 (8221/10501) [0.775, 0.791] |
| Model | Metric | Group | Overall | Normal | Mild | Moderate | Severe |
|---|---|---|---|---|---|---|---|
| Binary | Sensitivity | Overall | 0.633 (3948/6239) [0.621, 0.645] | 0.583 (74/127) [0.497, 0.668] | 0.551 (252/457) [0.506, 0.597] | 0.573 (1123/1960) [0.551, 0.595] | 0.676 (2499/3695) [0.661, 0.691] |
| H | 0.539 (2384/4427) [0.524, 0.553] | 0.581 (72/124) [0.494, 0.668] | 0.529 (220/416) [0.481, 0.577] | 0.523 (894/1710) [0.499, 0.547] | 0.550 (1198/2177) [0.529, 0.571] | ||
| O/A | 0.870 (1444/1660) [0.854, 0.886] | 1.000 (2/2) [1.000, 1.000] | 1.000 (5/5) [1.000, 1.000] | 0.920 (207/225) [0.885, 0.955] | 0.861 (1230/1428) [0.843, 0.879] | ||
| C/A | 0.790 (120/152 [0.725, 0.854] | 0.000 (0/1) [0.000, 0.000] | 0.750 (27/36) [0.609, 0.892] | 0.880 (22/25) [0.753, 1.000] | 0.789 (71/90) [0.705, 0.873] | ||
| PPV | 0.695 (3948/5681) [0.683, 0.707] | 0.260 (74/285) [0.209, 0.311] | 0.475 (252/531) [0.432, 0.517] | 0.674 (1123/1666) [0.652, 0.697] | 0.781 (2499/3199) [0.767, 0.796] | ||
| FP/patient | Overall | 39.0 [30.5, 47.6] | 23.4 [18.4, 28.5] | 39.9 [35.4, 44.3] | 36.1 [32.3, 40.0] | 52.8 [39.1, 66.4] | |
| In-sleep | 26.0 [18.6, 33.4] | 17.2 [13.1, 21.4] | 28.0 [23.7, 32.3] | 25.9 [22.2, 29.5] | 31.0 [18.7, 43.4] | ||
| Multiclass | Sensitivity | Overall | 0.622 (3883/6239) [0.610, 0.634] | 0.543 (69/127) [0.457, 0.630] | 0.490 (224/457) [0.444, 0.536] | 0.557 (1092/1960) [0.535, 0.579] | 0.676 (2498/3695) [0.661, 0.691] |
| H | 0.525 (2323/4427) [0.510, 0.539] | 0.540 (67/124) [0.453, 0.628] | 0.476 (198/416) [0.428, 0.524] | 0.508 (868/1710) [0.484, 0.531] | 0.547 (1190/2177) [0.526, 0.568] | ||
| O/A | 0.869 (1443/1660) [0.853, 0.886] | 0.500 (1/2) [0.000, 1.000] | 1.000 (5/5) [1.000, 1.000] | 0.898 (202/225) [0.858, 0.937] | 0.865 (1235/1428) [0.847, 0.883] | ||
| C/A | 0.770 (117/152) [0.703, 0.837] | 1.000 (1/1) [1.000, 1.000] | 0.583 (21/36) [0.422, 0.744] | 0.880 (22/25) [0.753, 1.000] | 0.811 (73/90) [0.730, 0.892] | ||
| PPV | 0.695 (3883/5585) [0.683, 0.707] | 0.199 (69/347) [0.157, 0.241] | 0.489 (224/458) [0.443, 0.535] | 0.673 (1092/1622) [0.650, 0.696] | 0.791 (2498/3158) [0.777, 0.805] | ||
| FP/patient | Overall | 38.5 [29.9, 47.0] | 30.9 [23.7, 38.1] | 33.4 [29.3, 37.5] | 35.3 [30.3, 40.2] | 50.1 [37.0, 63.2] | |
| In-sleep | 25.3 [17.7, 32.8] | 22.2 [16.2, 28.3] | 24.1 [20.4, 27.9] | 24.7 [20.0, 29.3] | 28.7 [16.6, 40.8] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, J.W.; Kim, D.H.; Koo, D.L.; Park, Y.; Nam, H.; Lee, J.H.; Kim, H.J.; Hong, S.-N.; Jang, G.; Lim, S.; et al. Automated Detection of Sleep Apnea-Hypopnea Events Based on 60 GHz Frequency-Modulated Continuous-Wave Radar Using Convolutional Recurrent Neural Networks: A Preliminary Report of a Prospective Cohort Study. Sensors 2022, 22, 7177. https://doi.org/10.3390/s22197177
Choi JW, Kim DH, Koo DL, Park Y, Nam H, Lee JH, Kim HJ, Hong S-N, Jang G, Lim S, et al. Automated Detection of Sleep Apnea-Hypopnea Events Based on 60 GHz Frequency-Modulated Continuous-Wave Radar Using Convolutional Recurrent Neural Networks: A Preliminary Report of a Prospective Cohort Study. Sensors. 2022; 22(19):7177. https://doi.org/10.3390/s22197177
Chicago/Turabian StyleChoi, Jae Won, Dong Hyun Kim, Dae Lim Koo, Yangmi Park, Hyunwoo Nam, Ji Hyun Lee, Hyo Jin Kim, Seung-No Hong, Gwangsoo Jang, Sungmook Lim, and et al. 2022. "Automated Detection of Sleep Apnea-Hypopnea Events Based on 60 GHz Frequency-Modulated Continuous-Wave Radar Using Convolutional Recurrent Neural Networks: A Preliminary Report of a Prospective Cohort Study" Sensors 22, no. 19: 7177. https://doi.org/10.3390/s22197177
APA StyleChoi, J. W., Kim, D. H., Koo, D. L., Park, Y., Nam, H., Lee, J. H., Kim, H. J., Hong, S.-N., Jang, G., Lim, S., & Kim, B. (2022). Automated Detection of Sleep Apnea-Hypopnea Events Based on 60 GHz Frequency-Modulated Continuous-Wave Radar Using Convolutional Recurrent Neural Networks: A Preliminary Report of a Prospective Cohort Study. Sensors, 22(19), 7177. https://doi.org/10.3390/s22197177





